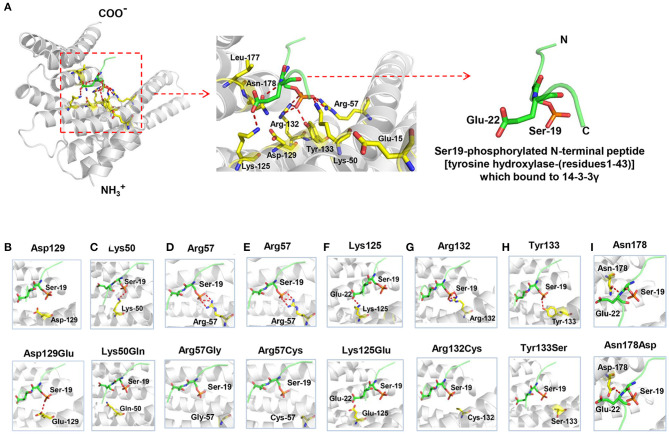
Figure 2.
Crystal structure of 14-3-3γ (PDB: 4J6S) and hydrogen bond changes of the mutants. (A) Left: monomeric 14-3-3γ is shown as gray ribbons, and the phosphopeptide ligand is shown as a green stick. Middle: close-up view of the binding groove and the side chains of the residues where mutations occurred. Right: Ser19-phosphorylated N-terminal peptide [tyrosine hydroxylase-(residues1-43)], which bound to 14-3-3γ. (B) Substitution of aspartate with glutamate at residue 129 (Asp129Glu) led to a new hydrogen bond with phosphopeptide ligand. (C) Substitution of lysine with glutamate at residue 50 (Lys50Gln) destroyed the original hydrogen bond with phosphopeptide ligand. (D) Substitution of arginine with glycine at residue 57 (Arg57Gly) destroyed the original hydrogen bond with phosphopeptide ligand. (E) Substitution of arginine with cysteine at residue 57 (Arg57Cys) destroyed the original hydrogen bond with phosphopeptide ligand. (F) Substitution of lysine with glutamate at residue 125 (Lys125Glu) destroyed the original hydrogen bond with phosphopeptide ligand. (G) Substitution of arginine with cysteine at residue 132 (Arg132Cys) destroyed the three original hydrogen bonds with phosphopeptide ligand. (H) Substitution of tyrosine with serine at residue 133 (Tyr133Ser) destroyed the original hydrogen bond with phosphopeptide ligand. (I) Substitution of asparagine with aspartate at residue 178 (Asn178Asp) formed a similar hydrogen bond with phosphopeptide ligand.