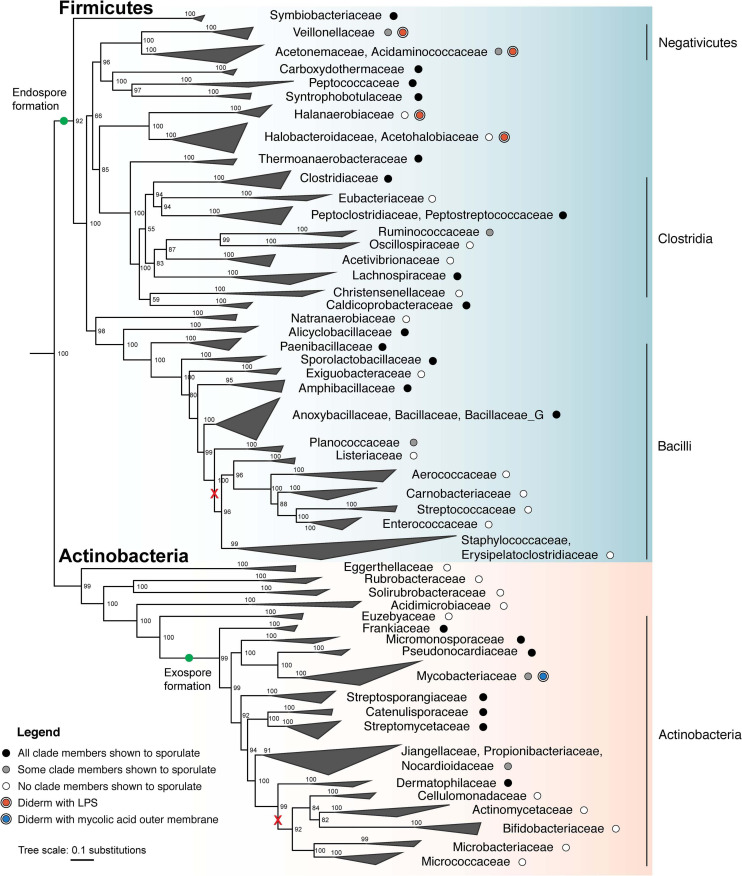
FIGURE 3.
Maximum likelihood phylogeny of sporulating and non-sporulating Firmicutes and Actinobacteria. Tree was constructed using an alignment of 120 single-copy marker gene sequences from several hundred representative genomes from each phylum using GTDB-Tk (Parks et al., 2018; Chaumeil et al., 2019). Whole genome phylogeny was determined using the concatenated marker gene alignment and IQ-TREE (v. 2.0.3), with the substitution model LG + G4 and 1000 ultrafast bootstraps (Minh et al., 2020). The tree was subsequently down-sampled and collapsed to show major families. Several representative Cyanobacteria genomes served as an outgroup. Tree was visualized using ggtree (Yu, 2020). Green dots indicate that endospore formation was likely present in the last universal ancestor of all Firmicutes, whereas exospore formation appears to have evolved after phylum differentiation. Black and gray dots represent the demonstrated ability to sporulate in all members and some members, respectively, whereas white dots represent the lack of sporulation ability. Major losses involving multiple families are shown (red x). Clades which include diderm bacteria are indicated for possession of either an LPS-containing OM (red dots) or a mycolic acid-containing OM (blue dot). Classes, as defined by the GTDB, are labeled on the right.