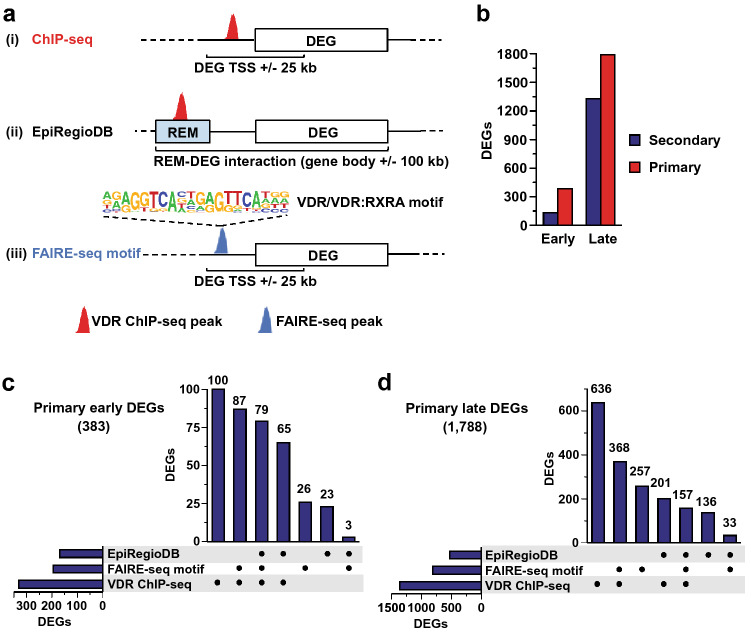
Figure 1.
Identification of direct VDR target genes. Three different approaches have been used to predict primary vitamin D target genes from early or late 1,25(OH)2D3-responding genes measured with RNA-seq11 (a). The first two identified direct target genes using VDR ChIP-seq peaks that were (i) located in a window of ± 25 kb around the TSS of genes or ii) overlapping a regulatory element (REM) in the EpiRegio database that was linked to a DEG. The third (iii) used FAIRE-seq peaks with a predicted VDR binding motif that are located in a window of ± 25 kb around the TSS of DEGs (b). The proportions of primary vitamin D target genes predicted by at least one of the three methods are shown for both early and late DEGs. The numbers of primary VDR target genes predicted by one, two, or all three of the approaches are depicted in upset plots for early (2.5 or 4 h; c) and late (24 h; d) DEGs after 100 nM 1,25(OH)2D3 treatment of THP-1 cells11,15.