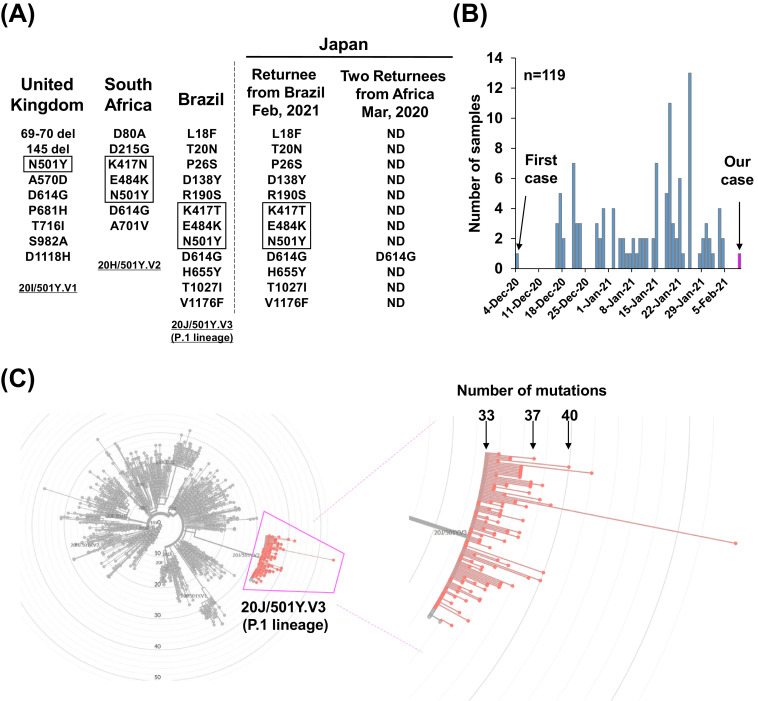
Fig. 2.
SARS-CoV-2 spike protein mutation in emerging lineages and global distribution. (A) Mutations in the SARS-CoV-2 spike protein. (Left) Amino acid changes identified in the emerging strains reported in the United Kingdom, South Africa, and Brazil. (Right) Results of the current analysis: the patient who returned from Brazil on February 2021 had the same mutation as 20 J/501Y.V3 (P.1 lineage); the two patients who returned from Africa on March 2020 had only a D614G mutation in the spike protein. The mutations highlighted by boxes indicate those in the receptor binding domain. ND, not detected. (B) The number of 20 J/501Y.V3 (P.1 lineage) strains deposited in GISAID by February 14, 2021. The first case was identified on December 4, 2020 and our case on February 10, 2021. (C) A total of 119 sequencing data were analyzed on Nextclade. (Left) Radial phylogenetic tree showing the location of 20 J/501Y.V3 (P.1 lineage). (Right) Magnified view of boxed area showing the P.1 lineage. The total numbers of mutations denoted are with respect to the SARS-CoV-2 strain from Wuhan, China.