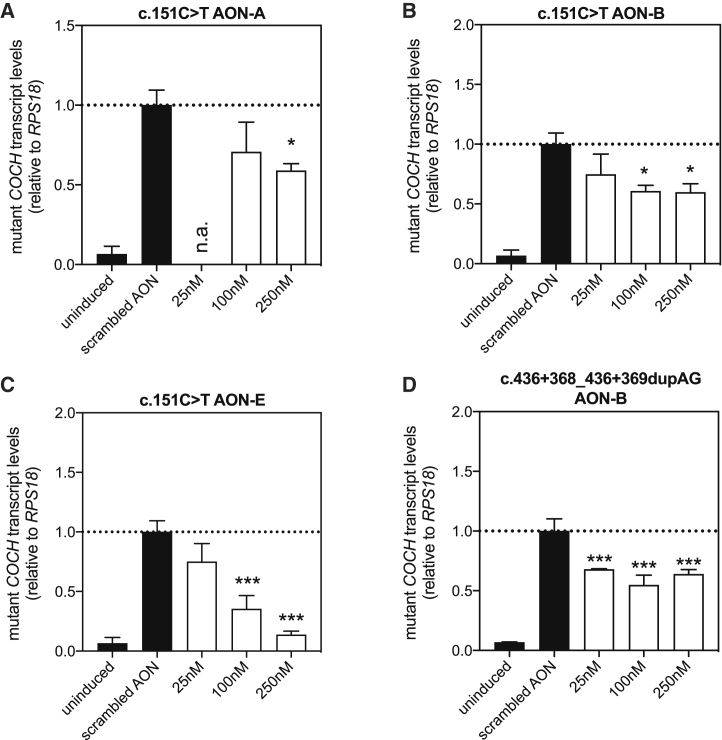
Figure 3.
Identified candidate AONs induce a significant decrease in mutant COCH transcript levels
(A–D) To confirm the effect of previously identified candidate AONs, c.151C>T AON-A (A), c.151C>T AON-B (B), c151C>T AON-E (C), and c.436+368_436+369dupAG AON-B (D) were investigated at two different doses. (A) c.151C>T AON-A results in significant decrease in mutant COCH transcripts at 250 nM, but not at 100 nM. (B) c.151C>T AON-B was able to induce a significant decrease in mutant COCH transcripts at both 100 nM and 250 nM, but no differences between the two doses were observed. (C) c.151C>T AON-E decreased the level of mutant COCH transcripts in a statistically significant and dose-dependent manner. At a concentration of 250 nM, the number of COCH transcripts was reduced to 20% of those in cells treated with a scrambled control AON. (D) Transfection of c.436+368_436+369dupAG AON-B resulted in a significant decrease of mutant COCH transcripts, without statistically relevant differences between the two concentrations. All four AONs had a gapmer design with wings of 2′-O-methyl-RNA bases flanking the central PS-DNA core. AONs were transfected at a dose leading to the indicated concentration in the well and investigated for their effect on transcript levels 24 h after transfection. Data are expressed as mean ± SD of 3 replicate transfections, normalized to the expression of RPS18 and displayed as the fold change compared to scrambled control AON-treated cells. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, one-way ANOVA with Tukey’s post-test.