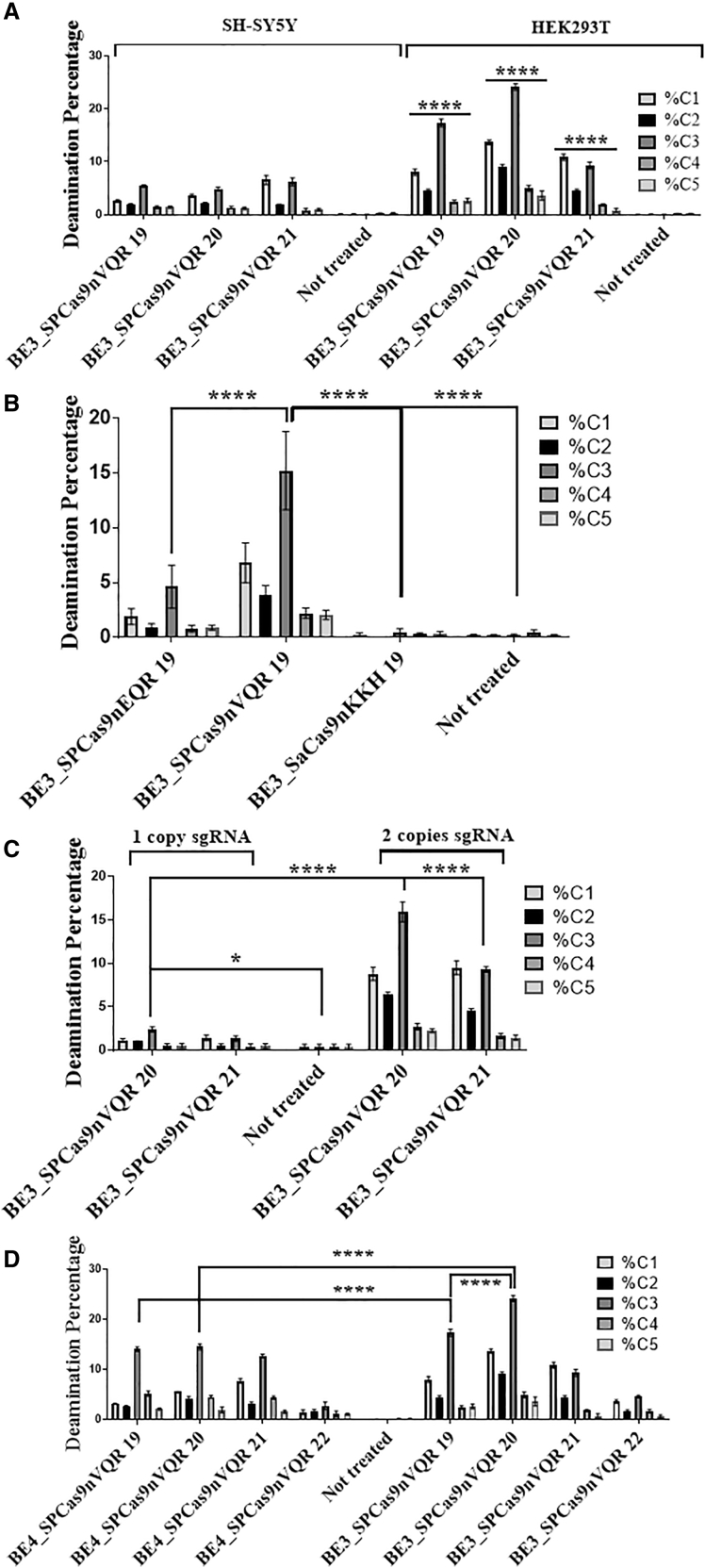
Figure 3.
Percentages of CDA produced by various enzymes and sgRNAs
Plasmids coding for the various base editors and for one or two sgRNAs were transfected in HEK293T and SH-SY5Y cells. The number of nucleotides targeted by the sgRNAs is indicated after the names of the enzymes. DNA was extracted 3 days post-transfection, APP exon 16 was PCR amplified and sent for deep sequencing. (A) Significant differences between SH-SY5Y and HEK2093T editing efficacy are shown with BE3_SpCas9nVQR. Transfected with two copies of sgRNA. The figure illustrates the mean ± SEM (n = 3). (B) BE3_SpCas9nEQR, BE3_SpCas9nVQR, and BE3_SaCas9nKKH enzymes were tested in HEK293T cells. They were transfected with two copies of sgRNA. The figure illustrates the mean ± SEM (n = 4). (C) The comparison between the use of one copy sgRNA versus two copies during base editing transfection in HEK293T cells is demonstrated. The figure illustrates the mean ± SEM (n = 3). (D) The BE4_SpCas9nVQR and BE3_SpCas9nVQR enzymes were tested in HEK293T cells. The figure illustrates the mean ± SEM (n = 3). SH-SY5Y results available in complementary results Figures S1 and S2. Statistical test: two-way ANOVA Tukey’s multiple comparisons test, (A) n = 3, (B) n = 4, (C) n = 3, and (D) n = 3. p value style: ∗p < 0.0332, ∗∗∗∗p < 0.0001.