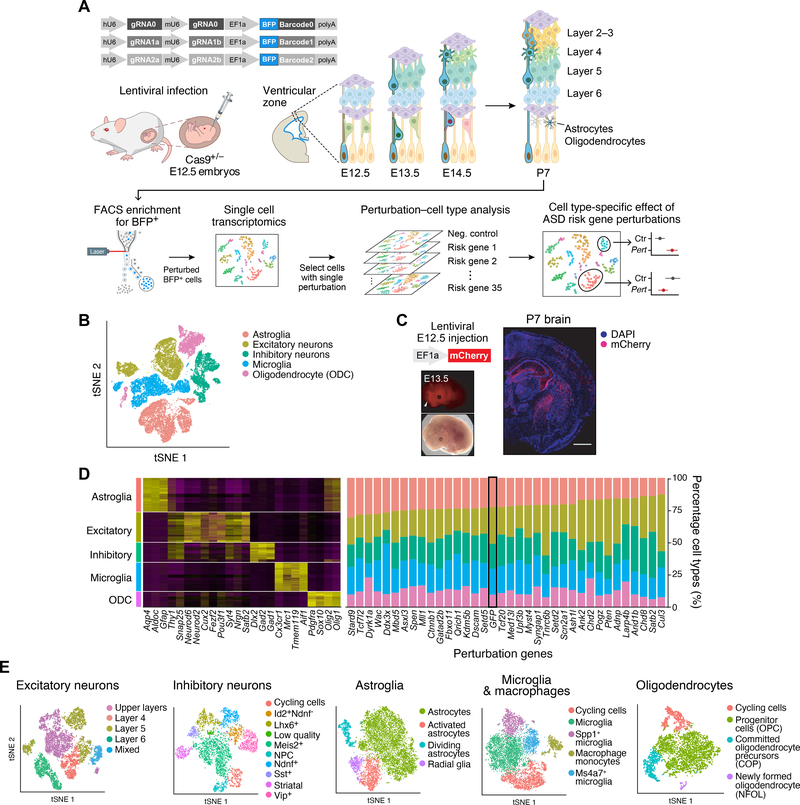
Figure 1. In vivo Perturb-Seq to investigate functions of a panel of ASD/ND risk genes harboring de novo variants.
(A) Schematics of the in vivo Perturb-Seq platform, which introduces mutations in individual genes in utero at E12.5, followed by transcriptomic profiling of the cellular progeny of these perturbed cells at P7 via single-cell RNA sequencing (scRNA-seq). (B) tSNE of five major cell populations identified in the Perturb-Seq cells. (C) In vivo Perturb-Seq lentiviral vector carrying an mCherry reporter drives detectable expression within 24h, and can sparsely infect brain cells across many brain regions. Scale bar is 1000μm. (D) Cell-type analysis of in vivo Perturb-Seq of ASD/ND de novo risk genes. Canonical marker genes were used to identify major cell clusters (left), and cell-type distribution in each perturbation group (right). Negative control (GFP) is highlighted by a black rectangle. (E) tSNEs showing the subclusters of each of the five major cell types, identified by re-clustering each cell type separately.