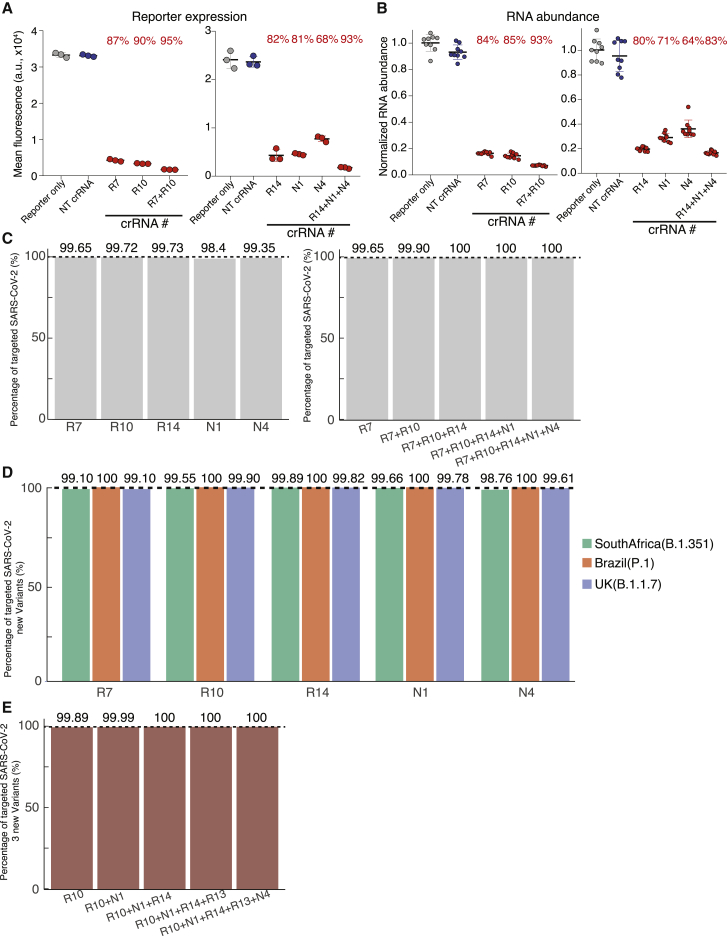
Figure 4.
Optimized crRNAs targeting SARS-CoV-2 sequences using CRISPR-PAC-MAN
(A and B) Repression effects of the optimized minipool of crRNAs (crRNA-MP5) on SARS-CoV-2 GFP reporter (A) and RNA abundance (B).
(A) GFP expression as measured by flow cytometry. p values for each group are included in Table S3. Lines indicate means; error bars indicate SDs (n = 3 independent biological replicates).
(B) mRNA abundance as measured by quantitative real-time PCR. Relative RNA abundance is calculated by normalizing to the reporter-only sample. p values for each group are included in Table S3. Lines indicate means; error bars indicate SDs. The data represent 3 independent biological experiments performed in technical replicates (n = 9).
(C) Bar graphs showing the percentage of genomes of SARS-CoV-2 isolates targeted by individual or multiple crRNAs in the optimized crRNA minipool.
(D and E) Bar graphs showing the percentage of genomes of 3 new SARS-CoV-2 variants targeted by individual (D) or multiple crRNAs (E) in the optimized crRNA minipool.
See also Figures S2 and S3 and Tables S3 and S4.