Fig. 11.
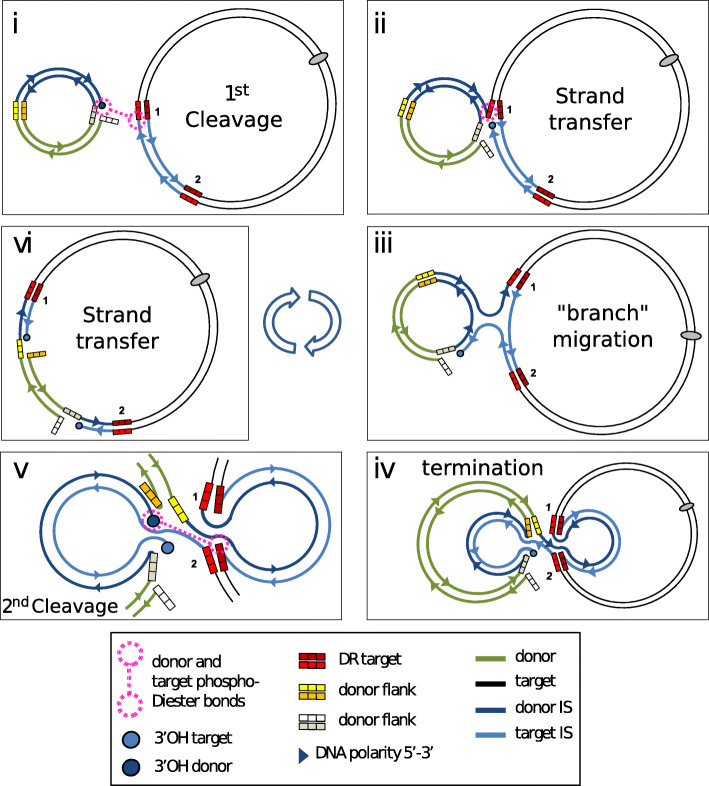
A model for IS26-mediated conservative targeted integration. i) Two IS ends from different IS copies in separate replicons are synapsed intermolecularly in the same transpososome, one end is cleaved to generate a 3’OH (shown as a dark blue circle) leaving a 5′ and on the flank (3 white boxes). This attacks the end of the second IS in the transpososome (shown as two dotted circles joined by a dotted line). ii) strand transfer would then couple the donor and target replicons via the target IS flank (3 bright red squares) leaving a 3’OH on the target IS (light blue circle). iii) strand migration can then occur in which one strand of the door IS and one strand of the target IS invade their partners. iv) following exchange of the entire partner strands, only a single physical strand cleavage would have occurred leaving a single single-strand break (three white squares). v) a second strand cleavage at the distal end of the donor IS occurs (dark blue circle) leaving its free 5′ flank (three orange squares). The 3’OH then attacks the distal target IS end (shown as two dotted circles joined by a dotted line). vi) strand transfer then generates a cointegrate with single-strand nicks at each end on opposite strands (white and orange squares) which could then be repaired. Note that the cointegrate retains the original flanking repeats of the target IS (three bright red and three dark red squares)