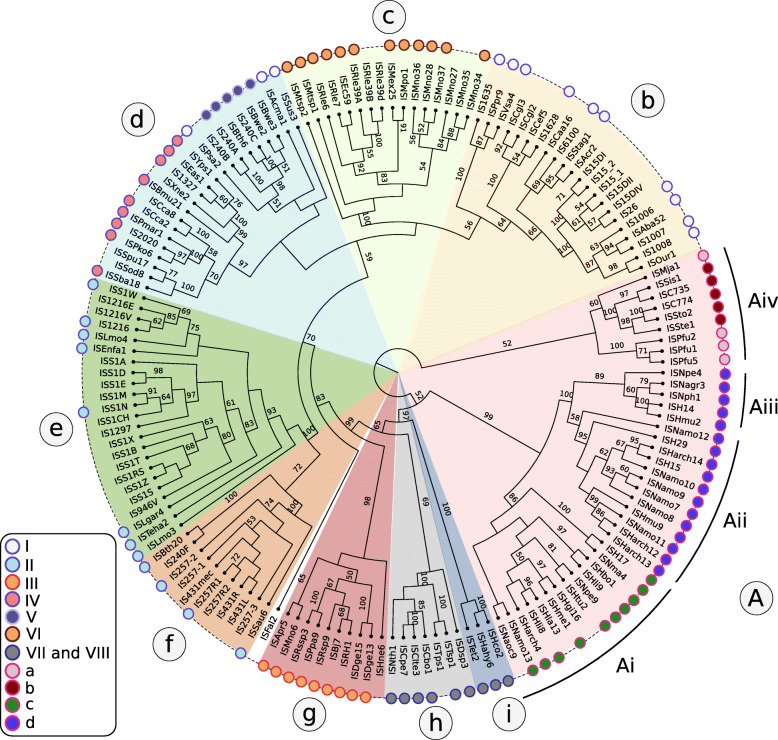
Fig. 3.
A dendrogram of IS6 family members.: A dendrogram of IS6 family members. The figure shows 11 major clades. The surrounding colored circles and the insert indicate the clades identified by [40]. The insert shows the correspondence between the clades from Harmer and Hall and those defined here. Clades: A: composed almost entirely of archea; Ai: (n = 12) is composed of diverse Halobacterial species (Halohasta, Haloferax, Natrinema, Natrialba, Halogeometricum, Natronomonas, Natronococcus, and Haloarcula); Aii: (n = 12) is composed uniquely of Halobacterial Euryarchaeota; Aiii: (n = 5) is composed entirely of Halobacterial Euryarchaeota (Haloarcula, Halomicrobium, Natronomonas, Natronobacterium, Natrinema); Aiv: (n = 9) which includes both Euryarchaeota and Crenarchaeota; b: (n = 16) Actinobacteria, Alpha-, Beta-, and Gamma-proteobacteria; c: (n = 14) Alphaproteobacteria: Rhizobiaceae and Methylobacteriaceae); d: (n = 24) (Alpha-, Beta-, and Gamma-proteobacteria, Firmicutes, Cyanobacteria, Acidobacteria and Bacteroidetes); e: (n = 23) is composed mainly of IS from Lactococcus, a single Leuconostoc and other bacilli (Lysteria, Enterococcus); f: (n = 11) largely Staphylococci with 2 B. thuringiensis; g: (n = 10) is heterogenous (Alpha proteobacteria: Methylobacterium, Paracoccus, Roseovarius, Rhizobium, Bradyrhizobium; Deinococci and Halobacteria); h: (n = 5) composed entirely of Firmicutes (Natranaerobius, Clostridium and Thermoanaerobacter); i: (n = 3) is composed of Halanaerobia and Thermoanaerobacter. TnpA protein sequences retrieved from ISfinder curated data set were aligned with MAFFT 7.309, and their best-fit evolutionary models were predicted with ProTest 3.2.4. A maximum likelihood tree was reconstructed with RaxML 8.2.9 using a bootstrap value of 1000. The final tree was visualized in FigTree 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree) and edited with Inkscape 0.92.4 (http://www.inkscape.org)