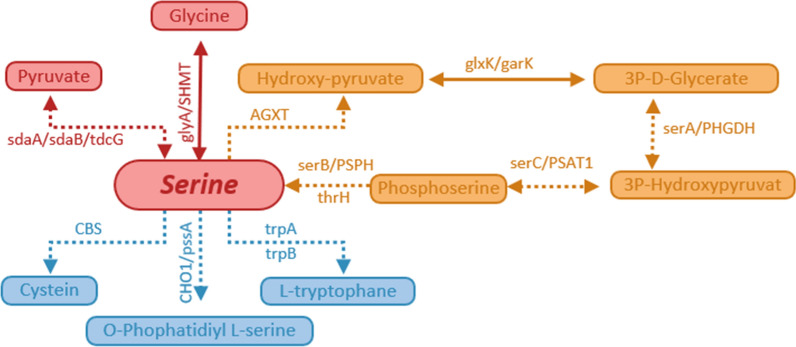
Fig. 10.
Glycine, serine, and threonine metabolism pathways of L. gasseri HHuMIN D. Solid arrows indicate that the metabolism pathways of glycine, serine, and threonine are effective and dashed arrows indicate invalid: sdaA/sdaB/tdcG L-serine dehydratase, glyA/SHMT glycine hydroxymethyltransferase, AGXT alanine-glyoxylate transaminase, glxK/garK glycerate 2-kinase, serA/PHGDH D-3-phosphoglycerate dehydrogenase, serC/PSAT1 phosphoserine aminotransferase, serB/PSPH phosphoserine phosphatase, thrH/phosphoserine homoserine phosphotransferase, CBS cystathionine beta-synthase, CHO1/pssA CDP-diacylglycerol–-serine O-phosphatidyltransferase, trpA tryptophan synthase alpha chain; and trpB, tryptophan synthase beta chain. Whole genome sequencing of L. gasseri HHuMIN D was performed by Chunlab, Inc. (Seoul, Korea), which used PacBio Sequel Systems (Pacific Biosciences, Menlo Park, CA, USA) and analyzed using CLgenomics™ and EZBioCloud Apps programs (Chunlab, Seoul, Korea). Biosynthetic pathways of glycine, serine, and threonine were analyzed by matching with the KEGG pathway using complete genome sequencing of L. gasseri HHuMIN D via CL Genomics™ (ChunLab, Seoul, South Korea, 2004). The activities of related genes and substances produced can be inferred through the biosynthetic pathways provided by KEGG