Figure 4.
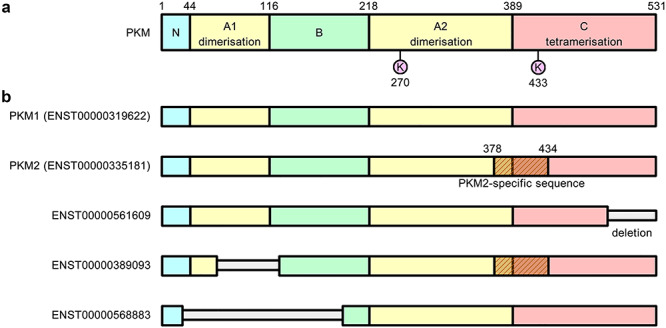
Alternative splice isoforms of PKM. Homology modelling and structure alignment can reveal functionally important sites and identify functionally significant deletions that occur in different PKM isoforms [62]. A, The template structure for PKM consists of four domains. The A-domain participates in the formation of dimers and the C-domain mediates the interactions between dimers that allow them to form tetramers. The active site (K270) and FBP binding site (K433) are shown. B, The alternatively spliced forms of PKM reveal large deletions corresponding to the ADP binding site in isoforms ENST00000389093 and ENST00000568883, which may impede dimerisation. In TGCA KIRC datasets, these transcripts are associated with unfavourable survival.
