Figure 2.
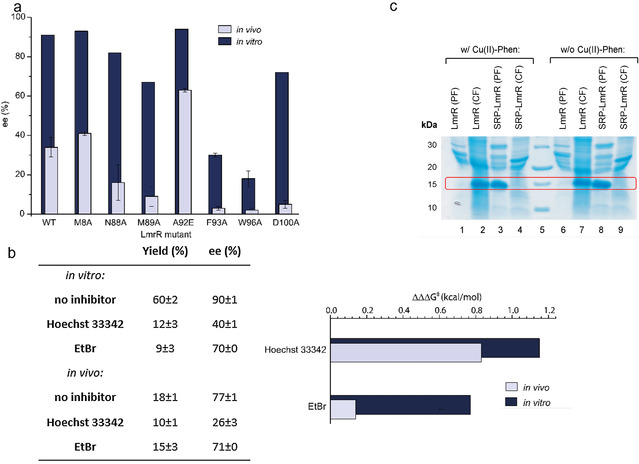
a) Enantioselectivity of the Friedel–Crafts alkylation reaction of 1 with 2 a, catalyzed by isolated CuII‐Phen/LmrR artificial enzyme mutants (dark blue) and CuII‐Phen/LmrR artificial enzyme mutants in whole E. coli cells (light blue). b) Left: Effect of addition of Hoechst 33342 and ethidium bromide (EtBr; 4 equivalents compared to CuII‐phen) on the yield and enantioselectivity of the reaction of 1 with 2 b, catalyzed by isolated CuII‐Phen/LmrR artificial metalloenyzmes and in whole cells. Values are given as the average of independent duplicate experiments, each performed in duplo. Errors are given as standard deviations. Right: Difference in enantioselectivity in the reaction of 1 with 2 b catalyzed by isolated CuII‐Phen/LmrR artificial metalloenzymes (dark blue bars) and in whole cells (light blue bars upon addition of inhibitors, compared to w/o inhibitor). A larger bar signifies a larger effect of the inhibitor on the enantioselectivity. Enantioselectivity differences are represented as ΔΔΔG ǂ, which is calculated using ΔΔΔG ǂ=ΔΔG ǂ (w/o inhibitor)−ΔΔG ǂ (w inhibitor) and ΔΔG ǂ=RTln(er), in which er is the enantiomeric ratio: % major enantiomer/% minor enantiomer. c) SDS PAGE of a E. coli cell fractionation experiment to determine protein localization for LmrR and SRP‐LmrR with and without CuII‐Phen. PF=periplasmic fraction; CF=cytoplasmic fraction.
