Table 7 .
Performance of the CTMC algorithm adding slices obtained from DrugBank.
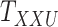
|
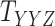
|
# of Slices | Runtime (s) | AUC | F1 | Sensitivity | Specificity | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |||
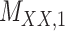
|
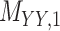
|
2 | 0.154 | 0.013 | 0.664 | 0.072 | 0.184 | 0.110 | 0.164 | 0.085 | 0.997 | 0.011 |
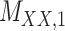 ,
, 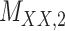
|
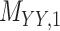
|
3 | 0.171 | 0.016 | 0.723 | 0.076 | 0.179 | 0.109 | 0.168 | 0.080 | 0.995 | 0.029 |
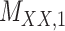 ,
, 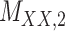
|
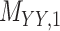 ,
, 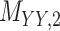
|
4 | 0.180 | 0.014 | 0.778 | 0.078 | 0.180 | 0.107 | 0.164 | 0.071 | 0.998 | 0.005 |
Metrics of results produced by the CTMC algorithm, stratified by the number of slices used in each coupled tensor. Here, 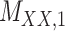 denotes binary drug–drug interaction matrix, and
denotes binary drug–drug interaction matrix, and 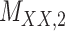 represents drug–drug similarity matrix with Morgan fingerprint scores. As for the targets,
represents drug–drug similarity matrix with Morgan fingerprint scores. As for the targets, 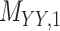 denotes binary target–target interaction matrix and
denotes binary target–target interaction matrix and 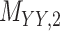 represents target–target similarity matrix as an inverse of Jukes–Cantor distance of amino acid sequences [12] (see Table 1 for more details on matrices
represents target–target similarity matrix as an inverse of Jukes–Cantor distance of amino acid sequences [12] (see Table 1 for more details on matrices 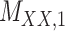 ,
, 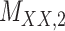 ,
, 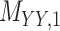 and
and 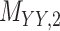 ).
).
