Figure 2.
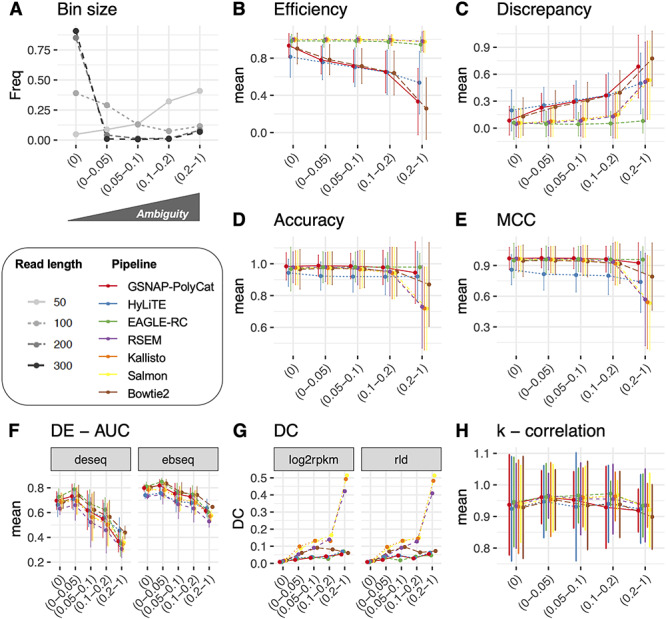
Homoeologous read ambiguity and consequences. (A) Given the specific sequencing read length (i.e. 50, 100, 200 and 300 bp), the homoeologous gene pairs from Gossypium were binned by ‘Ambiguity’ into five groups: 0, 0–0.05, 0.05–0.1, 0.1–0.2 and 0.2–1.0, the first of which indicates complete read assignment via SNP differentiation. The y-axis refers to the bin size of each gene group. These ‘Ambiguity’ bins were used to relate the performance of read assignment (B–E), DE (F), DC (G) and the analysis of node connectivity k (H). Error bars represent the standard deviation.
