Figure 1.
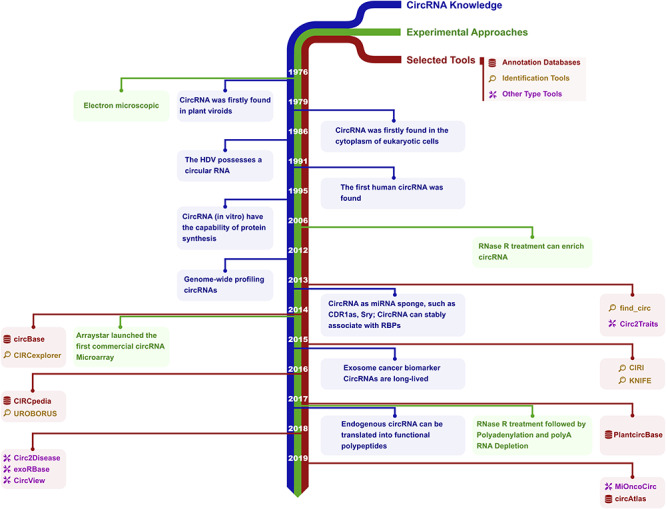
Historical timeline of circRNA research. The development of knowledge, experimental and computational tools of circRNA is illustrated. Blue, green and red marks are the circRNA discoveries related to biology, experimental approaches and representative bioinformatics tools, respectively. In 1976, circRNAs were identified as circular RNA genomes in plant viroids by electron microscopy [4]. With similar methods, circRNA was found in the cytoplasm of eukaryotic cells in 1979 [5] and in the hepatitis delta virus (HDV) in 1986 [170]. In 1991, the first human circRNA was identified [171]. In 1995, circRNA was shown to have the capability of protein synthesis in vitro [2]. In 2006, RNase R treatment was found to enrich circRNA [33]. By 2012, genome-wide profiling of circRNAs by RNA-Seq [8] was demonstrated. In 2013, circRNA was shown to act as a miRNA sponge with examples such as CDR1as [9] and Sry [172]. CircRNAs could stably associate with RNA binding proteins (RBPs), such as Argonaute protein (AGO) and RNA polymerase II [9, 31]. In 2014, Arraystar launched the first commercial circRNA microarray, and its expression was profiled by array [173]. In 2015, circRNAs were proposed as cancer biomarkers, and could be detected in the exosome [174]. CircRNAs also were shown to be long-lived [32]. In 2017, endogenous circRNA was shown to be translated into functional polypeptides [16, 166, 175]. New methods were developed for isolating highly pure circRNA using RNase R treatment followed by polyadenylation and polyA+ RNA depletion (RPAD) in 2017 [176].
