Figure 3.
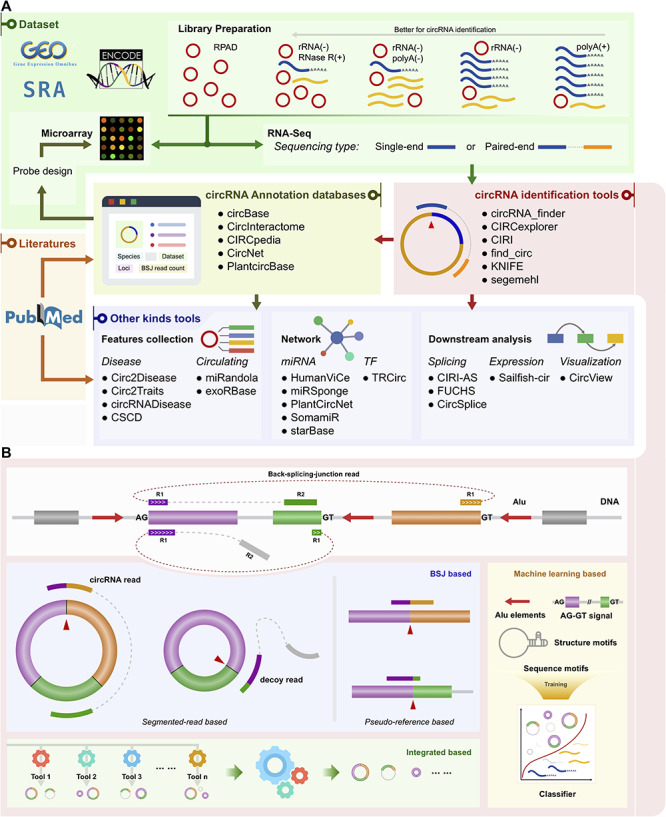
Schematic of classified circRNA bioinformatics tools. (A) Schematic of classified circRNA bioinformatics tools. Examples of tools are listed for each category. polyA(+): polyA selected; polyA(−): polyA depleted; rRNA(−): rRNA depleted; RNase R(+): treated with RNase R; RPAD: RNase R treatment followed by polyadenylation and polyA(+) RNA depletion. (B) Categories of circRNA identification tools. The top is a model of BSJ read mapping to the genome. R1 and R2 represent read1 and read2, respectively, in a paired-end RNA-Seq dataset. Three illustrations of categories are shown below the model. The categories are BSJ-based, integrated-based and machine learning-based. BSJ-based can be more finely divided into two sub-classes: segmented-read based and pseudo-reference based.
