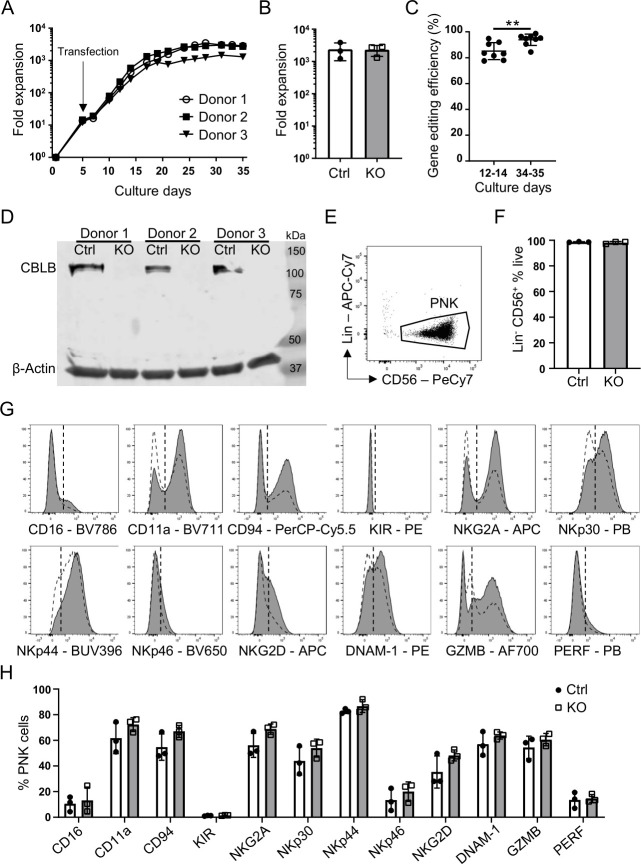
Figure 1.
Efficient CRISPR/Cas9-mediated CBLB gene editing and conserved NK cell identified in human PNK cells. (A) representative growth curves of CBLB KO PNK cell cultures. Cas9 mRNA and CBLB gRNA were introduced by transfection on day 5 (n=3). (B) Quantification of Ctrl or KO PNK cell expansion at day 35 of PNK culture, mean±SD (n=3). (C) CBLB gene editing efficiency using tide analysis summarizing eight individual experiments after transfection and on days 34–35 of PNK cell culture. Gene editing efficiency is determined as the frequency of nucleotide additions or deletions, mean±SD (n=8). (D) Western blot analysis of CBLB protein in day 35 Ctrl and KO PNK cells. β-actin serves as a loading control (n=3). (E) Representative dot plot of PNK cell gating. Indicated population was first gated of live single cells (not shown). Lins were CD3, CD14 and CD19. (F) Quantification of Lin− CD56+ cells in Ctrl and KO PNK cell cultures, mean±SD (n=3). (G) Representative histograms demonstrating the expression of indicated NK cell markers in Ctrl (white dashed) and KO PNK (gray) cells on day 35 of culture. KIR=CD158e1/e2 and CD158b1/b2, j. Vertical dotted line marks isotype control. (H) Quantification of the expression of indicated NK cell markers on Ctrl and KO cells, mean±SD (n=3). **P<0.01. CBLB, Casitas B-lineage lymphoma pro-oncogene-b; Ctrl, control; KIR, killer Ig-like receptor; KO, knockout; Lin, lineage marker; NK, natural killer; PNK, placental CD34+ cell-derived NK; PERF, perforin.