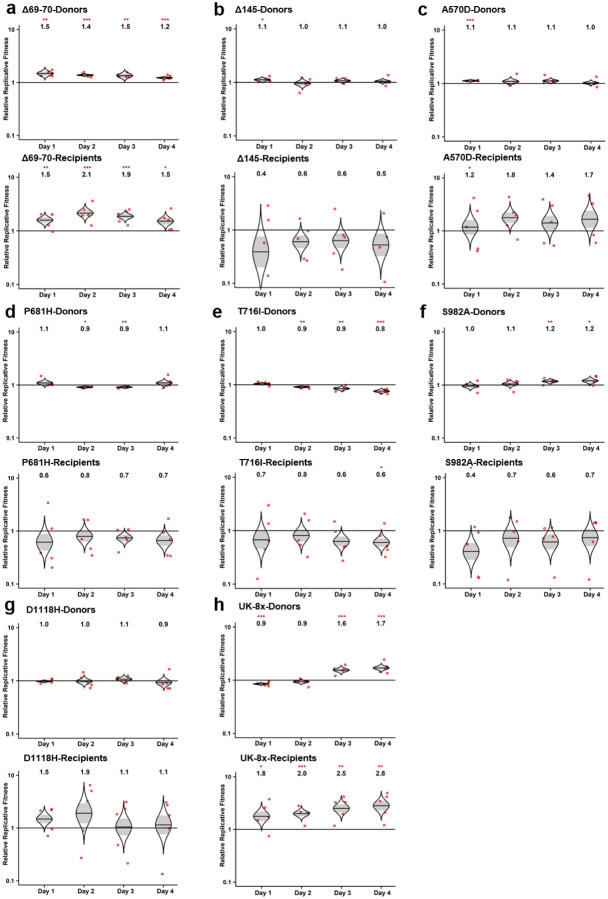
Extended Data Figure 2. The competition of other SARS-CoV-2 mutants with wt in hamsters.
a-h, Eight SARS-CoV-2 spike mutants: Δ69–70 (a), Δ145 (b), A570D (c), P681H (d), T716I (e), S982A (f), D1118H (g) and UK-8x (h) were mixed with wt virus at a ratio of approximately 1:1. The mixture was then inoculated intranasally into donor hamsters and transmitted to the recipient hamsters following the scheme in Figure 1a. The total titer for infection was 105 PFU per hamster. The ratios of the mutant:wt in the nasal washes of hamsters sampled 1–4 days after infection were estimated by Sanger sequencing. Red dots represent individual animals (n = 5), the horizontal lines in each catseye represent the mean, shaded regions represent standard error of the mean; y-axes use a log10 scale. Black numbers above each set of values (catseye) indicate the relative fitness estimates. P values are calculated for the group (strain) coefficient for each linear regression model. *p<0.05; **p<0.01; ***p<0.001.