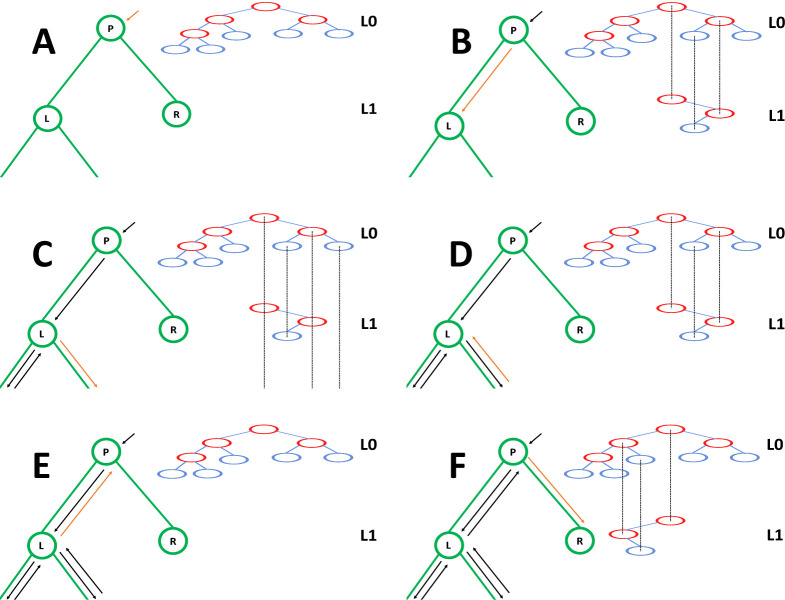
Fig 2. Example of multi-layer genome tree and its evolution.
We track the evolution of the multi-layer genome tree starting from the genome tree of Fig 1. Colors for the genome tree are the same as in Fig 1. In green, on the left side of each panel, we show an extract of the phylogenetic tree containing three nodes (“P” for parent, which in this example is the root of the phylogeny, and “L” and “R” for left and right node). “L” has further descendants, but we don’t show them here and only focus on this triplet of nodes as an example. The orange arrow along the phylogenetic tree shows the current step of the preorder traversal being considered by the given panel. Black arrows show past steps. Vertical dashed black lines in the multi-layer genome tree connect nodes that represent the same portions of the genome but that are in different layers. “L0” stands for “Layer 0” and “L1” for “Layer 1”. A At the phylogenetic root “P” we initialize the genome tree for layer 0. B As we move to child “L”, a new substitution is sampled (as in Fig 1) and 3 corresponding genome nodes are created in layer 1. These nodes correspond to the nodes in the original genome tree whose rate is affected by the new mutation. C As we traverse the subtree of the descendants of L, new nodes and mutations might be added in the layers below. D We are finished traversing the subtree of the descendants of L, and we return to L, at which point all nodes in layer below 1 have either been removed or have become irrelevant. E We return to P, at which point the genome tree nodes previously added layer 1 are also ignored or deleted. F We move from P to R, and in doing so new mutation events might be sampled and the corresponding genome nodes might be added to layer 1 (new genome tree nodes corresponding to 1 new substitution are shown in the new layer 1).