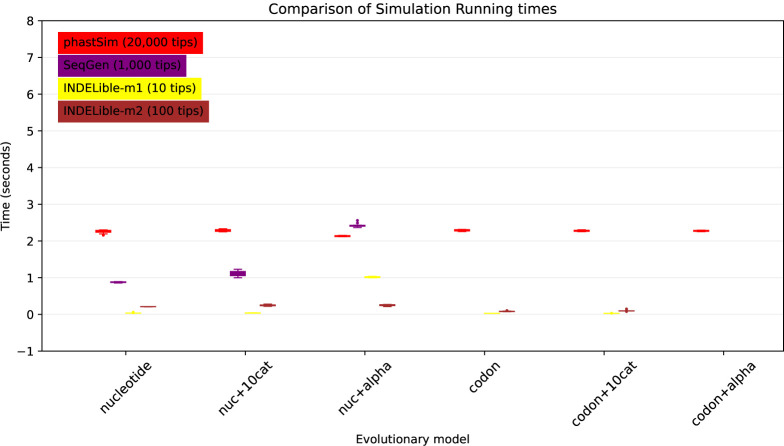
Fig 5. Comparison of running times of different simulators in a SARS-CoV-2 scenario using different evolutionary models.
On the Y axis we show the number of seconds it takes to perform simulations using different software. On the X axis is the model used for simulations: “nucleotide” is a nucleotide substitution model without variation; “nuc+10cat” is a nucleotide model with 10 rate categories; “nuc+alpha” is a nucleotide model with continuous variation in rate (each site has a distinct rate sampled from a Gamma distribution); “codon” represents a codon substitution model; “codon+10cat” represents a codon substitution model with 10 categories for ω; “codon+alpha” is a codon model with continuous rate variation in mutation rate and in ω (only allowed in phastSim). Each boxplot represents ten replicates. Seq-Gen does not allow codon models. Colors are as in Fig 3. To aid the visual comparison, we use trees of different sizes for different simulators: 10 tips for INDELible-m1; 100 tips for INDELible-m2; 1,000 tips for SEq-Gen; 20,000 tips for phastSim.