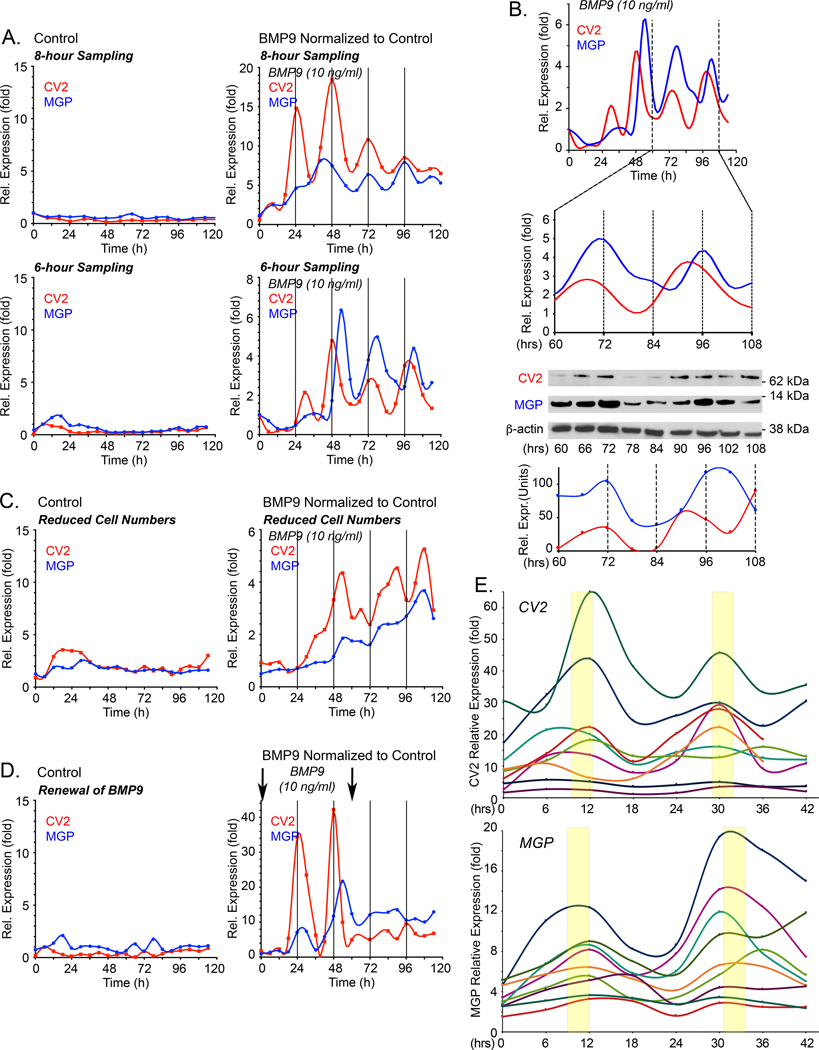
Figure 2: Temporal oscillations of CV2 and MGP transcripts in HPAECs in response to BMP9.
Expression profiles for MGP and CV2 under different conditions. In the following experiments, expression of MGP and CV2 was determined by qPCR, first normalized to GAPDH and then its own control. (A) HPAECs were treated with control medium (left panels) or BMP9 (10 ng/ml) (right panels), RNA was collected every 8 or 6 hours for up to 120 hours, and expression of CV2 and MGP was determined by qPCR. Adjusted R2 and p-values from non-linear polynomial regression were as follows: 8-hour collection: MGP: adjusted R2 0.841, p-value 7.63E-05; CV-2: adjusted R2 of 0.674, p-value 0.003; 6-hour collection: MGP: adjusted R2 0.499, p-value 0.005; CV-2: adjusted R2 0.406, p-value 0.027. For 8- and 6-hour collections, 3 and 5 replicate experiments were performed, respectively. (B) Corresponding CV2 and MGP protein levels from cells treated with BMP9 (10 ng/ml), between 60–108 hours (qPCR, immunoblot and image quantification) (representative of 3 replicate experiments). (C) HPAECs seeded at low density (6.25×103 cells/cm2), treated with control medium (left panel) or BMP9 (10 ng/ml) (right panel). RNA was collected every 6 hours for up to 120 hours, and expression of CV2 and MGP was determined by qPCR (duplicate experiments). (D) HPAECs were treated with control medium (left panel) or BMP9 (10 ng/ml) added at the time of plating and 60 hours later (arrows) (right panel). RNA was collected every 6 hours for up to 120 hours, and expression of CV2 and MGP was determined by qPCR (duplicate experiments). (E) HPAECs treated with BMP9 (10 ng/ml) for 42 hours; two early oscillations from 9 replicate experiment are shown.
Each qPCR time point represents the mean of 3 determinations, and each BMP9-treated time point is normalized to its own untreated control.