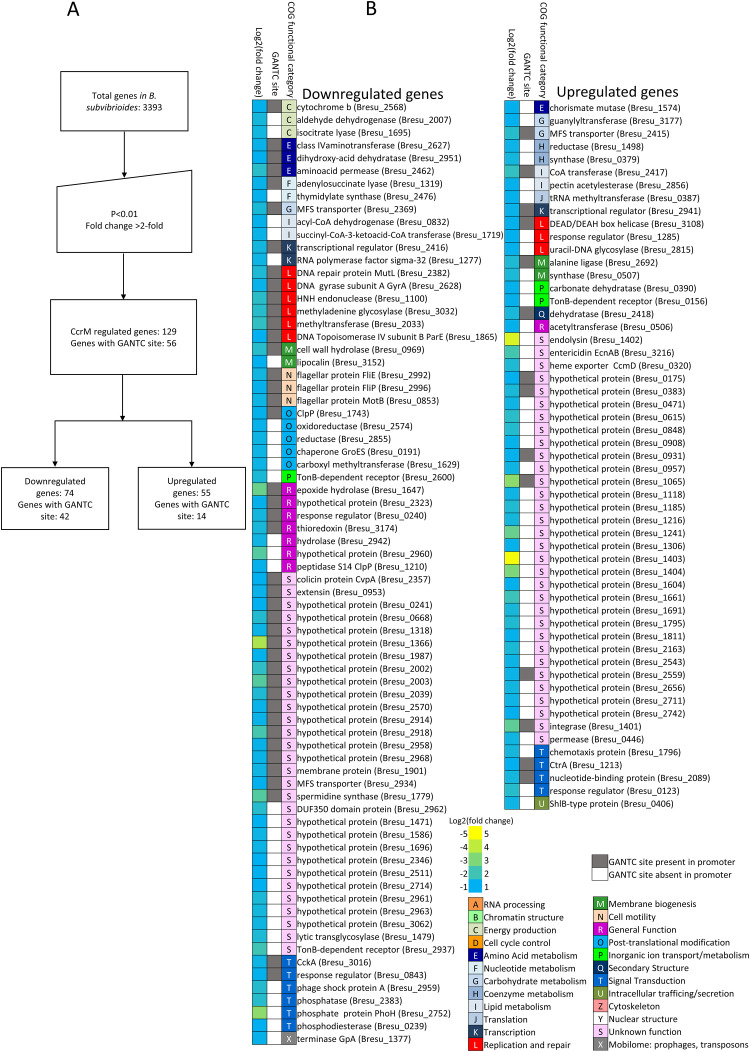
Fig 1. Genes misregulated in ccrM mutant compared to wild-type in B. subvibrioides.
A) Workflow showing the cutoffs used for defining misregulated genes in ccrM mutant. Using >2-fold and P<0.01 as cut offs, 129 genes were found misregulated compared to WT out of 3393 total genes in B. subvibrioides. Out of 129 misregulated genes, 56 of them had at least one GANTC site in their promoter suggesting potential direct regulation by CcrM. B) List showing genes downregulated (left) and upregulated (right) in the ccrM::pNPTS139 strain along with COG functional category. For both left and right, Column 1 shows the heat map of the magnitude of fold change in log2 scale. Column 2 shows if those genes have GANTC site within their promoter (grey—GANTC site present, white—GANTC site absent). Genes were clustered by COG functional category (Column 3).