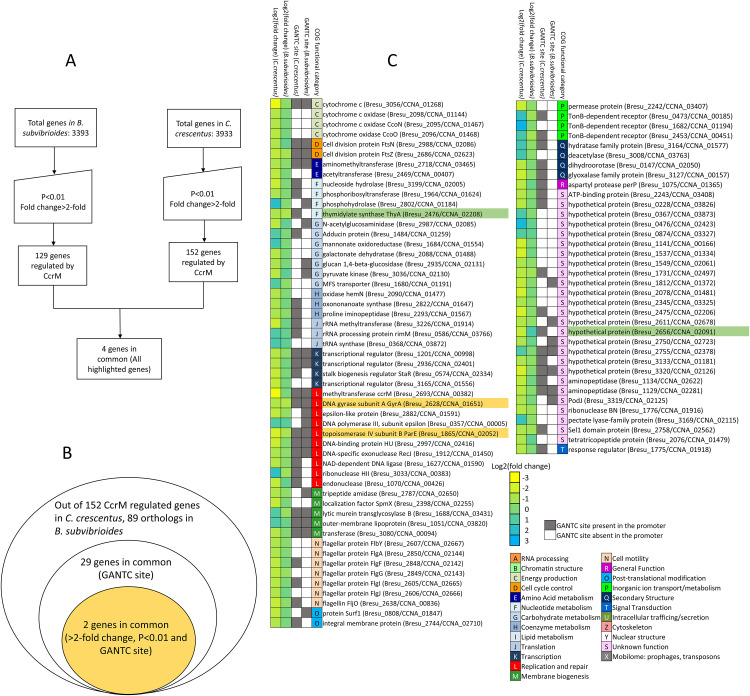
Fig 2. Common genes misregulated in ccrM mutant in B. subvibrioides and C. crescentus.
A) Workflow showing the cutoffs used for defining misregulated genes in ccrM mutant in both organisms. Using >2-fold and P<0.01 as cut offs, 129 genes and 152 genes were found misregulated in ccrM in B. subvibrioides and C. crescentus respectively. Only 4 genes were found in common. B) Concentric circle diagram showing common genes using different parameters. C. crescentus CcrM regulates 152 genes and B. subvibrioides has orthologs to 89 of those genes. Only 29 of the B. subvibrioides orthologs have GANTC sites in their promoter regions. Of those 29, only 2 genes showed significant transcriptional changes in a ccrM mutant strain (highlighted in orange). C) List of 89 B. subvibrioides orthologs to C. crescentus ccrM regulated genes sorted by COG functional category. Column 1 and 2 shows the heat map of the magnitude of fold change in log2 scale in the C. crescentus ccrM strain (data obtained from [23]) and B. subvibrioides ccrM::pNPTS139 strain respectively. Columns 3 and 4 show if those genes have GANTC site within their promoter in C. crescentus and B. subvibrioides respectively (grey—GANTC site present, white—GANTC site absent). Genes were clustered by COG functional category (Column 5). Orthologs that met the transcriptional change cutoffs are highlighted in green; orthologs that met the transcriptional change cutoffs and have a GANTC site are highlighted in orange.