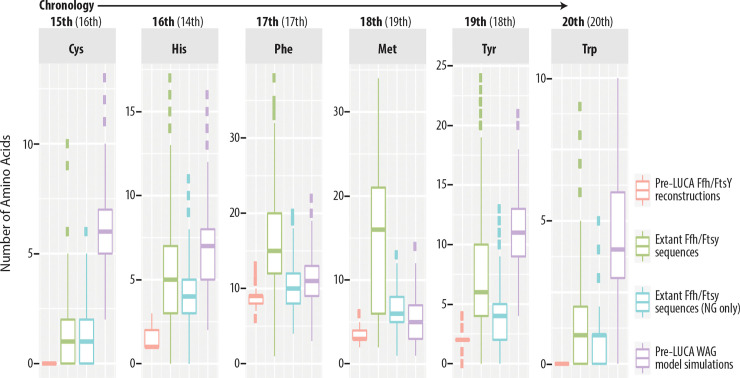
Fig 8. Paucity of late amino acids in ancestral Ffh/FtsY.
The chronological order of the addition of late amino acids to the genetic code is taken from Trifonov [52] and Jordan et al. [53] (the latter shown within parentheses). Distributions of amino acid counts from the ancestral Ffh/FtsY sequence (red) represent 100 sequence reconstructions for the pre-LUCA node using the 10% gap threshold in FastML. Distributions of amino acid counts are shown for extant SRP system proteins (green) or only the region containing the helical bundle and GTPase domains that aligns to the ancestral Ffh/FtsY sequence (blue). When analyzing extant proteins, we excluded FlhA and type III secretion system proteins from the analysis. In all cases, the inferred ancestral Ffh/FtsY sequences contain fewer late amino acids than extant homologs. Reconstructions based on simulated sequences derived from the WAG model (purple) demonstrate that the paucity of late amino acids in ancestral Ffh/FtsY is not an artifact of the ancestral sequence reconstruction methods or the background rate of amino acid usage across proteins in general.