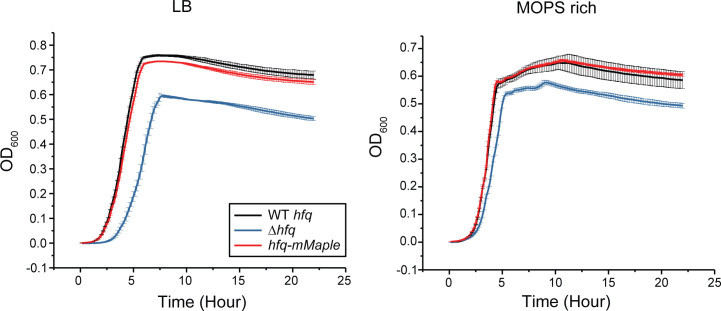
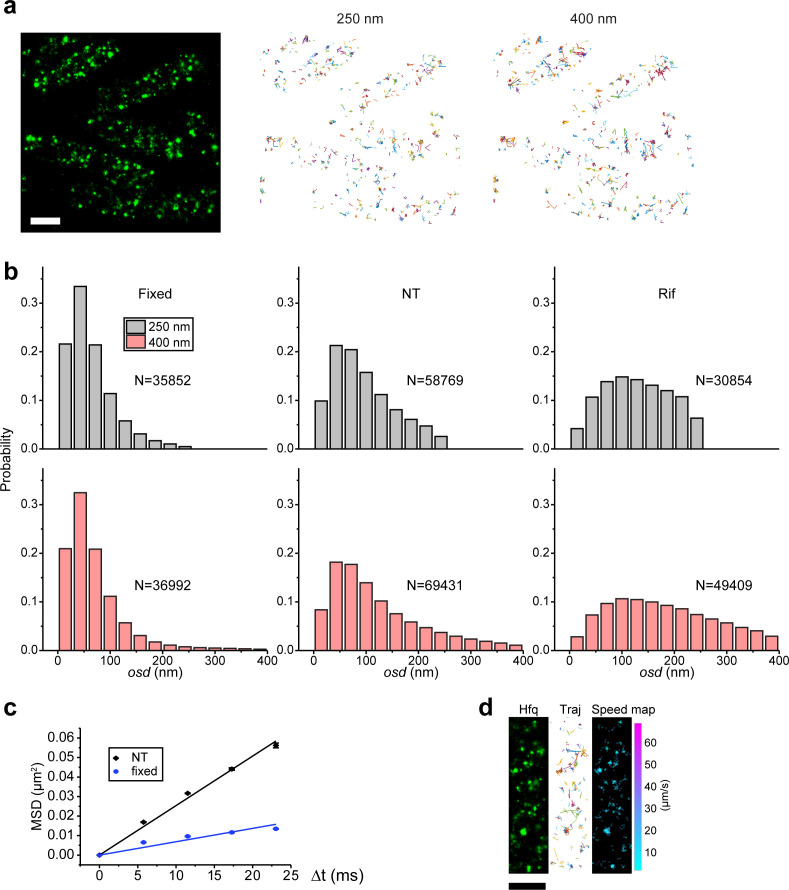
Figure 1. Diffusion and localization of Hfq during exponential growth.
(a) Schematic representation of Hfq with three RNA binding faces indicated. (b) A representative example of WT Hfq-mMaple3 in WT rne background in a single cell during exponential growth under no treatment (NT) condition. Nucleoid is stained with Hoechst in live cells. 2D reconstructed image of Hfq-mMaple3 is shown in the black background. Different diffusion trajectories from tracking algorithm are shown in different colors (‘Traj’). One-step displacement (osd) speed map (unit: µm/s) is shown as a scatter plot where different colors represent different speeds at each position, and the white curves represent the nucleoid regions detected by Hoechst staining. The scale bar represents 1 µm. (c) Enrichment of Hfq localization is calculated for cytoplasm, membrane, and nucleoid regions under NT condition. (d) Average osd speed of Hfq within the cytoplasm, membrane, and nucleoid regions under NT condition. Error bars in all plots represent the standard deviation (s.d.) from two experimental replicates, with each data set containing ~20,000 trajectories from ~80 cells.