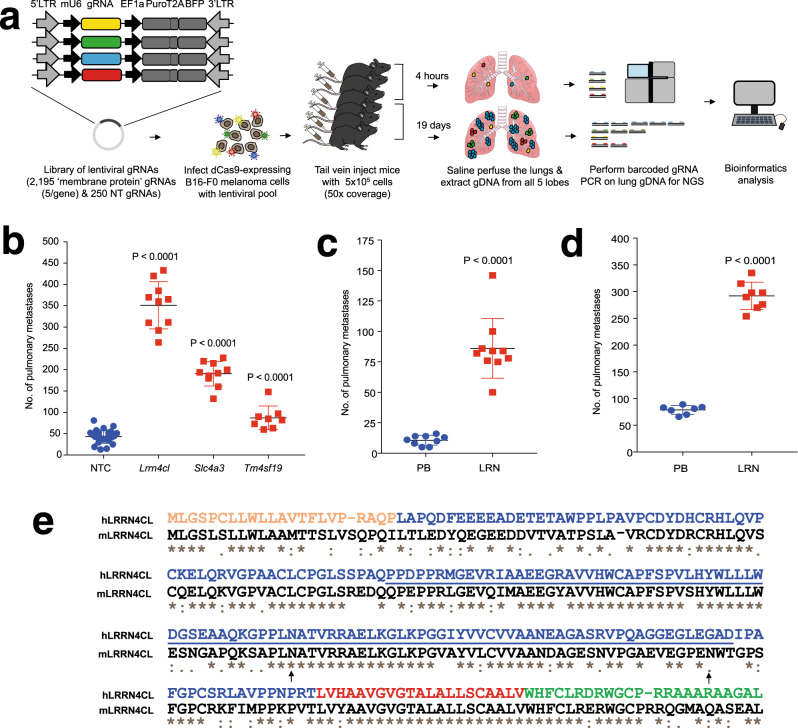
Fig. 1. Performing a CRISPRa screen in vivo to identify cell surface regulators of pulmonary metastatic colonisation.
a Graphic representation of the screen outline. b The number of metastatic colonies in the lungs of mice 10 days after being tail vein dosed with 2 × 105 B16-F0-dCas9 cells carrying gRNAs against the genes listed. NTC, non-targeting control. c The number of metastatic colonies in the lungs of mice 10 days after being tail vein dosed with 1 × 105 B16-F0 cells that have been stably transfected with a plasmid carrying the Lrrn4cl cDNA (LRN) or an empty vector (PB). d The number of metastatic colonies in the lungs of mice 10 days after being tail vein dosed with 4 × 105 B16-F0 cells that have been stably transfected with a vector carrying the Lrrn4cl cDNA (LRN) or an empty vector (PB). For (b-d), each symbol represents a mouse, the bars represent mean ± SD, 2 independent experiments performed (representative data from one experiment is shown) and statistics performed using a Mann–Whitney t test. e The human and mouse LRRN4CL protein sequences (ENSP00000325808 and ENSMUSP00000093976, respectively) were aligned CLUSTAL W (1.81) in Ensembl. Below each site amino acid of the alignment is a key denoting conserved sites (*), sites with conservative replacements (:), sites with semi-conservative replacements (.), and sites with non-conservative replacements (). The UniProtKB predicted location of the signal peptide (orange), extracellular domain (blue), transmembrane domain (red), cytoplasmic domain (green), FN3 domain (underlined) and N-linked glycosylation site at N132 (arrow) for human LRRN4CL are shown (glycosylation sites for the mouse also shown: arrows at N132 and N174).