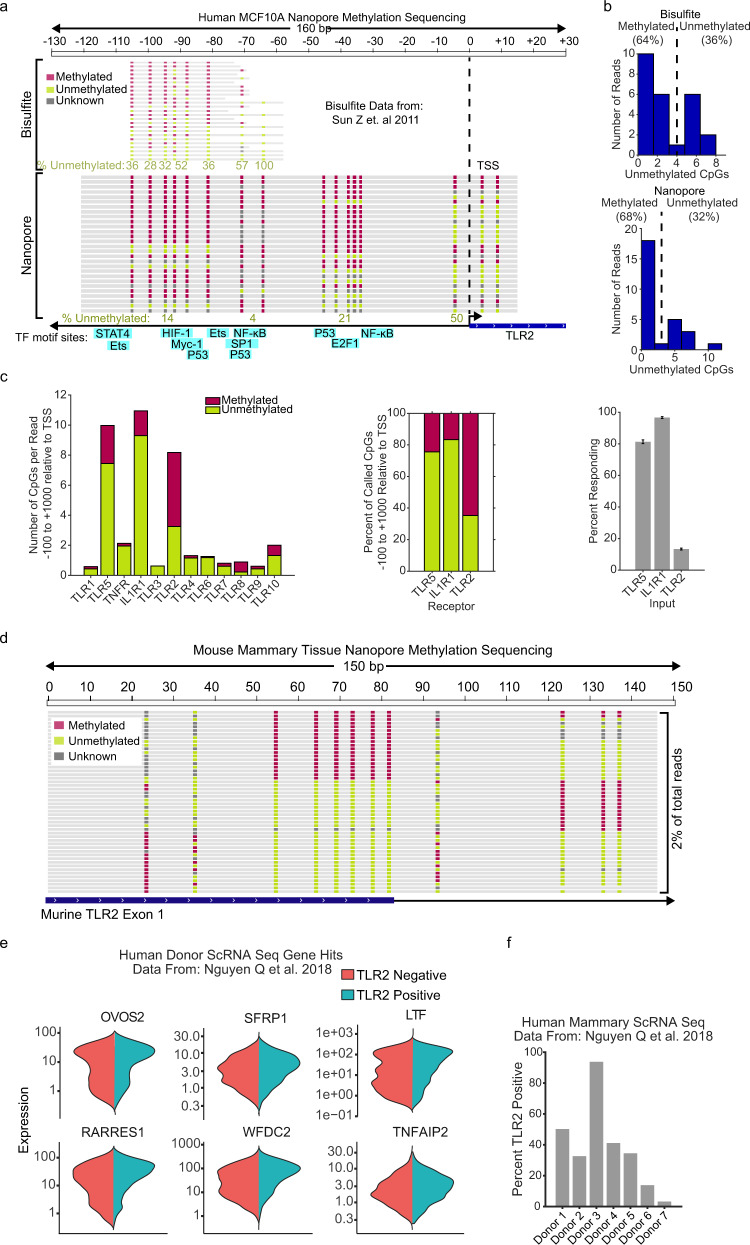
Fig. 5. TLR2 promoter methylation is bimodal.
a Bisulfite36 and nanopore sequencing data of 130 bp upstream of the TLR2 transcription start site (TSS) in MCF10A. Locations of transcription factor consensus binding sequence sites are indicated. b Histogram quantifications of the number of unmethylated CpGs per read between −110 and −60. c Nanopore methylation sequencing57 data of 100 bp upstream to 1000 bp downstream of TSS for indicated receptor. Percentage of unmethylated and methylated CpGs as well as percent responding cells as determined by p65 immunofluorescence following TLR5 (Flagellin), IL1R (IL-1B), and TLR2 (Pam) stimulation. d TLR2 locus targeted nanopore sequencing of genomic DNA isolated from primary mouse epithelial mammary cells. e Differentially expressed genes cosegregating with TLR2 negative and positive cells in publicly available single-cell RNA sequencing data from human breast epithelial tissues40. f Percent TLR2 positive cells from ScRNA sequencing data collected from breast tissue of seven female donors. Only cells expressing luminal expression markers were considered (see Methods section for details).