FIGURE 7.
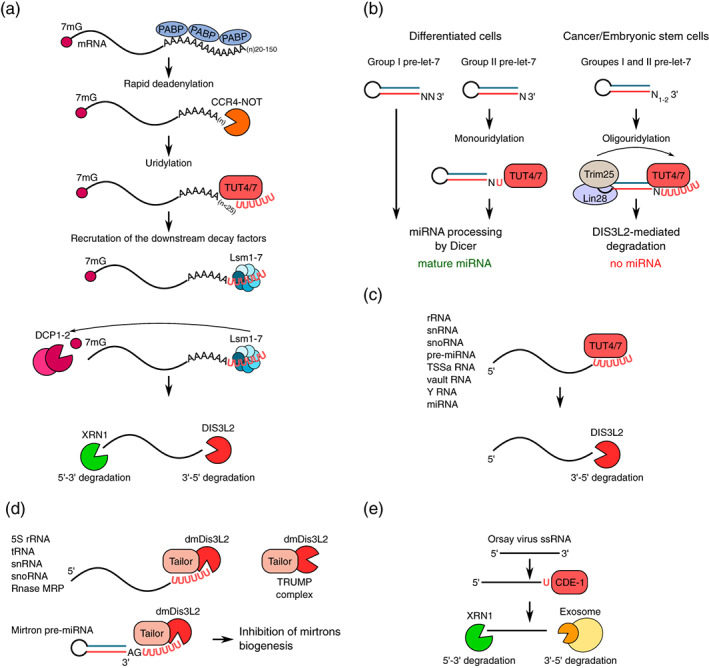
Selected uridylation‐dependent processes in humans, worms, and flies. (a) Uridylation‐mediated degradation of polyadenylated mRNAs in humans. After deadenylation, mRNAs with poly(A) tails that are shorter than 25 nt are uridylated by TUT4/7, leading to the recruitment of downstream RNA decay factors. The LSM1‐7 complex first interacts with the U‐tail and promotes mRNA decapping by the DCP1‐2 complex. mRNAs that are devoid of a cap are then degraded by the 5′–3′ exonuclease XRN1. Additionally, mRNA can be degraded by the exosome and/or DIS3L2 exonuclease in the 3′‐5′ direction. (b) Pre‐miRNA let‐7 uridylation by TUT4/7. Following their transcription and initial processing, the precursor miRNA (pre‐miRNA) are exported into the cytoplasm (not shown). In differentiated cells where Lin28 is absent, group II pre‐miRNAs that carry a one‐nucleotide 3′ overhang are monouridylated by TUT4/7, thus enabling further processing by Dicer. In cancer and embryonic stem cells, Lin28 binds both groups of let‐7 pre‐mRNAs and recruits TUT4/7. Dicer cannot process oligouridylated pre‐miRNAs, and they are degraded by the 3′‐5′ exonuclease DIS3L2. (c) Various RNA species that are transcribed by Pol I, Pol II, and Pol III can be targeted to the 3′–5′ exosome and/or DIS3L2 exonuclease degradation in a uridylation‐mediated manner. (d) In Drosophila, the decay of multiple RNAs relies on uridylation by Tailor, which forms a complex with the 3′–5′ exonuclease dmDis3L2 (a so‐called TRUMP complex). The stability of mirtrons is particularly controlled by uridylation. Mirtrons arise during splicing and lariat debranching of introns and carry AG at their 3′ ends. The 3′‐AG is uridylated by Tailor, which inhibits their biogenesis and leads to degradation by dmDis3L2. (e) Antiviral RNA uridylation in worms. Upon infection, Orsay virus RNAs are monouridylated by CDE‐1, which promotes their degradation by the 5′–3′ exonuclease XRN1 and 3′–5′ exonucleases of the exosome
