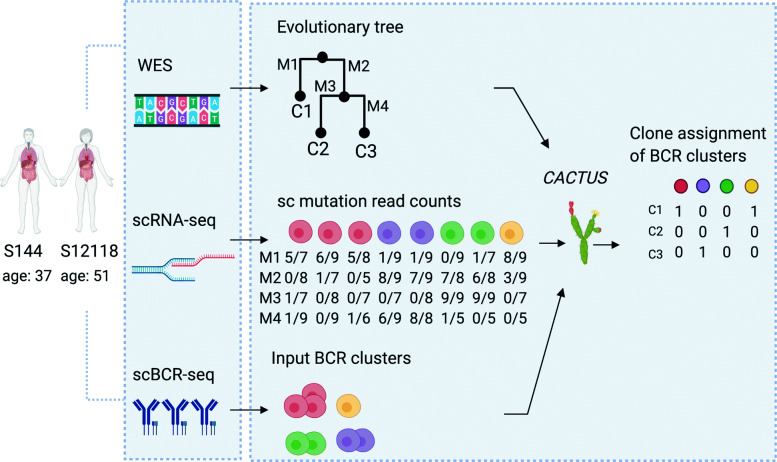
Fig. 1.
Overview of the patient data analysis and the CACTUS model. Whole-exome sequencing and single-cell sequencing of all transcripts, as well as single-cell sequencing of BCR, were performed on samples from two FL patients. Using WES, imperfect clonal evolution could be inferred and given as a prior to the model (C1, C2, …). From scRNA-seq, allele-specific transcript counts (mutated/total) were extracted at mutated positions (M1,M2, …). Input BCR clusters were defined as clusters of cells with identical BCR heavy chain sequences. The data of input tumor clones, mutation transcript counts, and given single-cell clusters (here, the BCR clusters) are combined in the CACTUS model for inference of the clonal assignment of the clusters. Both the input clone genotypes and clustering are considered potentially imperfect and are corrected during the inference using all available data. Image created with Biorender.com