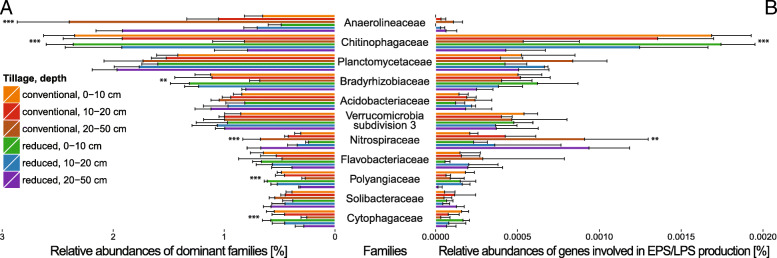
Fig. 1.
Dominant families and their potential to produce EPS and LPS. Comparison of (A) abundances of dominant bacterial families whose relative abundances exceeded 0.5%, with (B) their potential to produce EPS and LPS. Taxonomic assignment was performed against the National Center for Biotechnology Information Non-Redundant (NCBI-NR) protein sequences database. Functional genes were assigned using hidden Markov models (HMMs) obtained from the TIGRFAMs and Pfam databases, and then sequences derived from the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology database. Significant differences in the amount of annotated reads were determined by a multilevel model (n = 3). Detected influence is symbolized for depth (*) or interaction of tillage and depth (#), respectively. Significance levels are represented by the amount of symbols: 1 – p < 0.05, 2 – p < 0.01, 3 – p < 0.001. Error bars show standard deviation