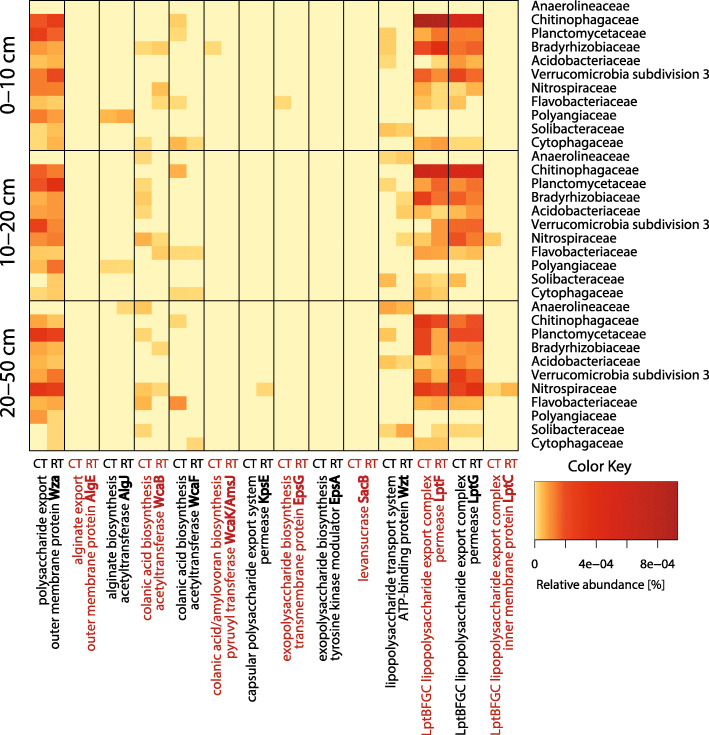
Fig. 3.
Genes encoding for proteins involved in EPS and LPS biosynthesis found in the dominant families. Heatmap representation of the mean relative numbers of genes encoding for proteins involved in EPS and LPS synthesis and excretion found in the dominant bacterial families whose abundance exceeded 0.5% in the samples from Frick taken at three depths. “CT” and “RT” on the x axis stand for “conventional tillage” and “reduced tillage”, respectively. Functional genes were assigned using hidden Markov models (HMMs) obtained from the TIGRFAMs and Pfam databases, and then sequences derived from the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology database. Taxonomic assignment was performed against the National Center for Biotechnology Information Non-Redundant (NCBI-NR) protein sequences database