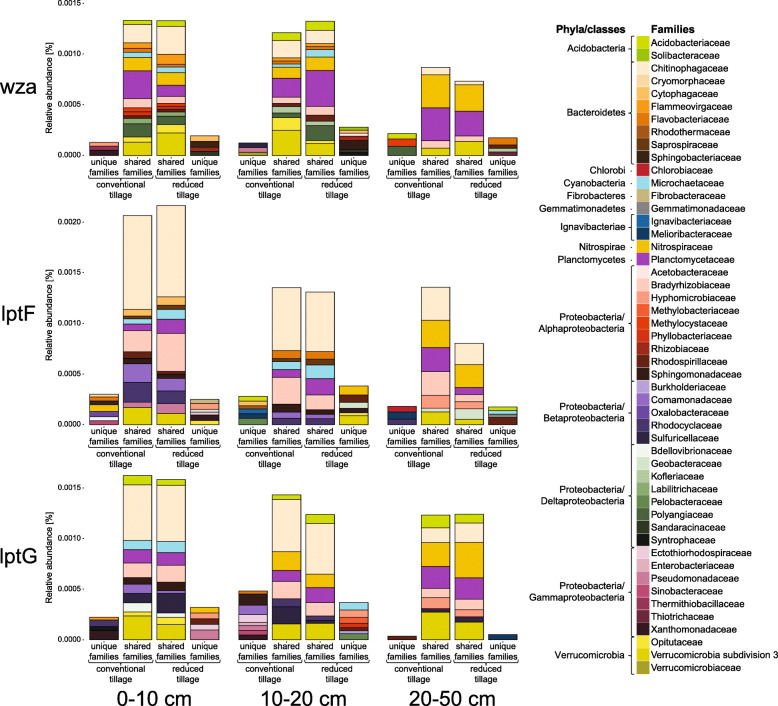
Fig. 4.
Taxonomic assignment of the wza, lptF and lptG genes. Mean relative abundances of the wza, lptF and lptG genes, encoding respectively for a polysaccharide outer membrane exporter and permeases of the LptBFGC LPS export complex, in the samples taken at three depths from plots under conventional and reduced tillage managements. Displayed are the distributions of gene copies among bacterial families listed on the right side of the graph. Only families found in at least two out of three replicates are presented. The color code is arranged according to the phylogenetic affiliation of the respective families. Compared are gene abundances in families harboring the respective genes uniquely under conventional or reduced tillage and families harboring the genes under both tillage managements. Functional genes were assigned using hidden Markov models (HMMs) obtained from the TIGRFAMs and Pfam databases, and then sequences derived from the Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology database. Taxonomic assignment was performed against the National Center for Biotechnology Information Non-Redundant (NCBI-NR) protein sequences database