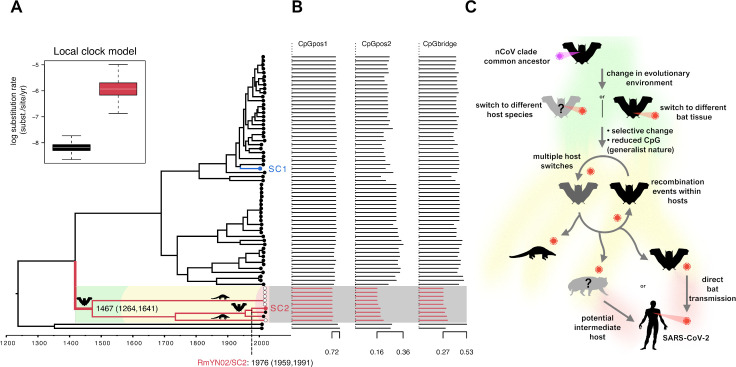
Fig 3.
(A) Bayesian phylogeny of a modified NRR2 region using different local clocks for the nCoV clade (red branches) and the rest of the phylogeny. All internal nodes’ posterior values are above 0.98. Viruses infecting bats are indicated by black circles, pangolins white circles, and SC1 and SC2 labelled in blue and red, respectively, at the tips of the tree. The inset summarises the substitution rate estimates on a natural log scale for the two-parameter local clock model with colours corresponding to the branches in the tree. The estimated date for the shared common ancestor of the nCoV clade (1467) and the RmYN02/SARS-CoV-2 divergence (1976) are shown with confidence intervals. (B) CpG relative representation for all dinucleotide frame positions (pos1: first and second codon positions; pos2: second and third codon positions; bridge: third codon position and first position of the next codon) is presented as SDUc values. (C) Schematic of our proposed evolutionary history of the nCoV clade and putative events leading to the emergence of SARS-CoV-2. The list of GenBank and GISAID accessions for the Sarbecovirus sequences used are provided in S4 Table. nCoV, new coronavirus; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; SC1, SARS-CoV-1; SC2, SARS-CoV-2; SDUc, corrected synonymous dinucleotide usage.