Fig. 2. Structural considerations of the identified mutations in human Na+/K+ ATPase α-subunits.
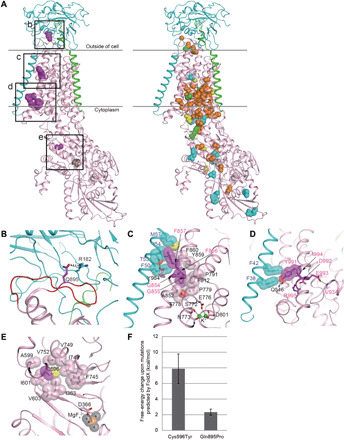
(A) The crystal structure of Na+/K+ ATPase, composed of α- (pink), β- (cyan), and γ-subunits (green), from Squalus acanthias (PDB code: 2ZXE) is shown with the residues (magenta spheres) corresponding to the identified variants in humans. The squares (b to e) correspond to the magnified areas (B to E), respectively. In the right structure, spheres indicate the residues corresponding to the previously reported variants associated with AHC (orange), RDP (cyan), both AHC and RDP (green), and other phenotypes (yellow). (B) The p.Arg182 (cyan) of the β-subunit interacting with p.Gln895 (magenta) in a loop of p.Asp882–Trp896 (red wire) of the α-subunit via hydrogen bonds (black dotted lines). The residue numbering is based on human Na+/K+ ATPase. (C) The interactions between α- and β-subunits around the p.Gly854 to Phe857 region of the α-subunit. The side chains of p.Phe856 and p.Phe857 and their interacting residues (β-subunit) are shown by sticks with translucent van der Waals spheres. The conformation of p.Met57 (β-subunit) in humans was modeled instead of leucine in the S. acanthias structure. The sulfur and oxygen atoms are shown in yellow and red, respectively. The residues interacting with K+ ions (green) are depicted as sticks. (D) The interactions between α and β subunits around the p.Tyr991-Arg995 region of the α-subunit. The p.Tyr991 and its interacting residues, p.Phe38 and p.Phe42 (β-subunit), are shown as sticks with translucent spheres. The conformation of p.Ile994 in humans was modeled instead of methionine in the S. acanthias structure. (E) A hydrophobic core involving p.Cys596 near the p.Asp366 phosphorylation site. The magnesium and fluorine atoms of a phosphate analog, MgF42−, are shown in orange and gray, respectively. (F) Free-energy changes upon the substitution of p.Cys596Tyr and p.Gln895Pro, as predicted by FoldX (31, 32).
