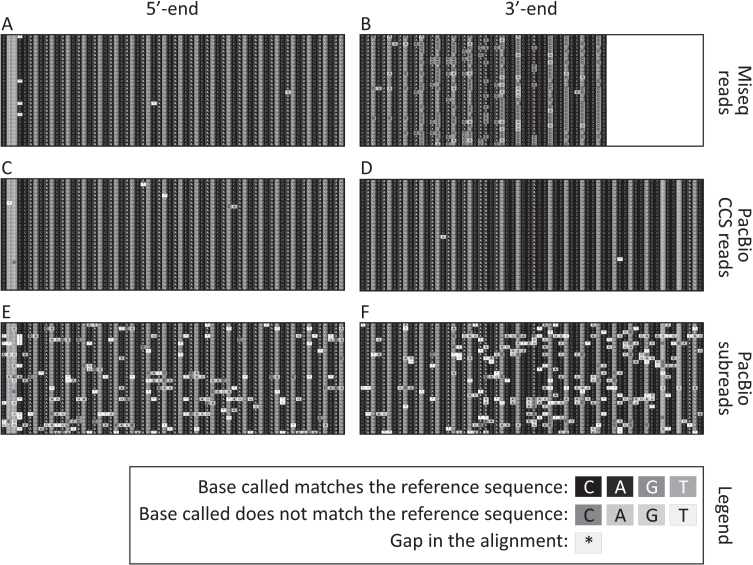
Fig. 2.
Representative sequence alignments of the 400 nt MiSeq reads (A and B), PacBio CCS reads (C and D) and PacBio subreads (E and F) uniquely aligned (i.e., reads not discarded post alignment) to a synthetic reference sequence with 115 CAGs. Alignments shown correspond to 30 sequencing reads obtained from the tail at weaning of the 20-week-old mouse with ∼110 CAGs. The part of the alignment shown corresponds to the four nucleotides in the immediate 5’–flank of the HTT CAG repeat, followed by the first 20 CAGs (A, C and E), as well as the last 7 CAGs followed by (CAACAG)1(CCGCCA)1(CCG)7(CCT)2 and the four nucleotides in the immediate 3’-flank of that sequence (B, D and F). Note that the last nucleotide sequenced for the sample with the 400 nt MiSeq reads end was the first C of the seventh CCG (B). The white box on the right-hand side of panel B represents the part of the PCR products containing 115 CAGs that could not be sequenced using 400 nt MiSeq reads.