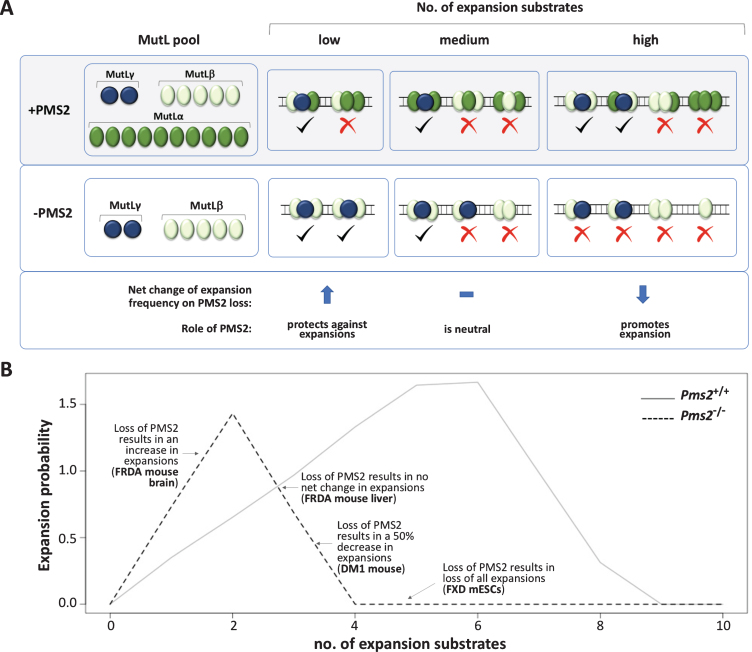
Fig. 2.
MutL competition model for the differential effect of the loss of PMS2 seen in different cell types or disease models. A simple mathematical model was developed for the competition between MutL proteins for binding to the expansion substrates. This model used the following assumptions: 1) Only a small proportion of the total cellular MutL is actually available for binding to the repeat; 2) Any one of the three MutLs can be recruited to a MutS-bound substrate; 3) Three MutLs (a MutL trimer) are required to bind productively to a substrate [123]; 4) The available MutL is distributed across all the substrates in proportion to their levels/binding affinity. 5) Only those MutL trimers that contain at least one MutLγ complex results in an expansion (indicated by a check mark); 6) Trimers that lack MutLγ or lesions that are not bound by at least three MutL complexes do not produce an expansion (indicated by a cross). A) Diagrammatic representation of the model showing MutL binding when the expansion substrates are present at different levels in the presence or absence of PMS2, with tick marks indicating outcomes that lead to expansions and the crosses those that do not. The number of available MutL complexes was set at MutLα= 10; MutLβ= 5 and MutLγ= 2, a ratio similar to that reported in mammalian cells [72]. When expansion substrate levels are low and PMS2 is present, not all MutLγ is bound, since PMS2 competes effectively for binding to the expansion substrate. As a result many MutL trimers formed lack MutLγ and their substrates are repaired without expansion. In the absence of PMS2, more MutLγ is able to bind and MutLβ contributes to the formation of additional MutL trimers required for MutLγ-generated expansions. As a result, a net increase in expansions is seen relative to cells with PMS2. At intermediate levels of substrate more MutLγ is able to bind and when PMS2 is absent, the residual MutLβ is sufficient for trimer formation at all MutLγ-bound sites. This results in no net change in the expansion frequency relative to cells with PMS2. However, at high levels of substrate, MutLβ becomes rate-limiting when PMS2 is absent, resulting in a net decrease in expansions. B) Graphical representation of the expansion probabilities across the range of substrate levels in the presence or absence of PMS2 based on the average of 1000 independent tests of the chances of binding of MutLα, MutLβ and MutLγ for each of the substrate levels. The python script used to generate the data upon which the graph is based is provided in the Supplementary Material. As in panel A, the number of available MutL complexes used was MutLα= 10; MutLβ= 5 and MutLγ= 2. However, as shown in the Supplementary Material similar results in terms of the range of effects of the loss of PMS2 are seen with wide range of different proportions of MutLα, MutLβ and MutLγ and with a wide range of absolute levels of total MutL.