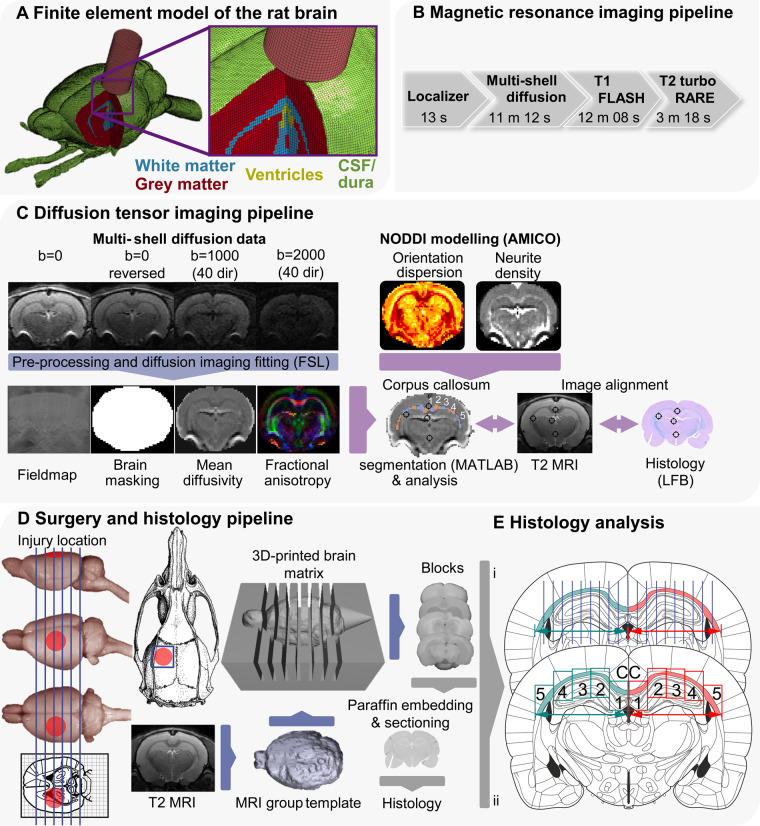
Figure 1.
Overview of methodology. (A) The FE model of the rat CCI. The image shows CSF (green), grey matter (red), white matter (blue), ventricles (yellow) and impactor (pink). The skull and dura are not shown. (B) MRI pipeline: diagram showing the acquisition protocol. (C) DTI pipeline: flow chart of diffusion MRI image analysis. Following the acquisition of scans, all files were converted from Bruker format to NIfTI. Post-processing was performed with FSL tools topup, bet and eddy correct before independent simultaneous diffusion and neurite orientation dispersion and density imaging fitting (AMICO). The last stage involved image alignment with T2 MRI, histology and vice versa. This alignment was based on anatomical landmarks identified in the histology staining, MRI, the Paxinos and Watson, and Waxholm rat brain atlases. The corpus callosum was manually outlined and automatically segmented. A representation of the five segments obtained across the corpus callosum in each hemisphere is presented and comparable to E(ii). (D) Surgery and histology pipeline: approximate location of craniotomy and impact is shown on the rat skull and brain. Animals were subjected to either 1 (n = 10) or 2 mm CCI (n = 11.) From T2 images, a grouped 3D template was derived, which was 3D printed with 2 mm intervals. Blocks were cut from a selection of animals (1 and 2 mm CCI: n = 6; naive/sham animals: n = 7) using the matrix and one block (4, containing the core of the contusion) was selected for paraffin embedding. From paraffinized blocks, 7 µm sections were cut and every fifth section collected on slides (three per slide), therefore covering roughly 100 µm. (E) Sections were stained and analysed according to the described protocols and segments of the corpus callosum analysed using FIJI and HALO (analysis approach i: LFB; ii: microglia, astrocytes and neurofilaments). These sections were aligned with the MRI data, based on the procedures described in C. Rat brain, skull and atlas images from Paxinos and Watson (2007) and the University of Wisconsin-Madison Brain collection (http://neurosciencelibrary.org/Specimens/rodentia/labrat/index.html).