Extended Data Figure 1. MK-SpikeSeq reliably measures absolute abundances across kingdoms.
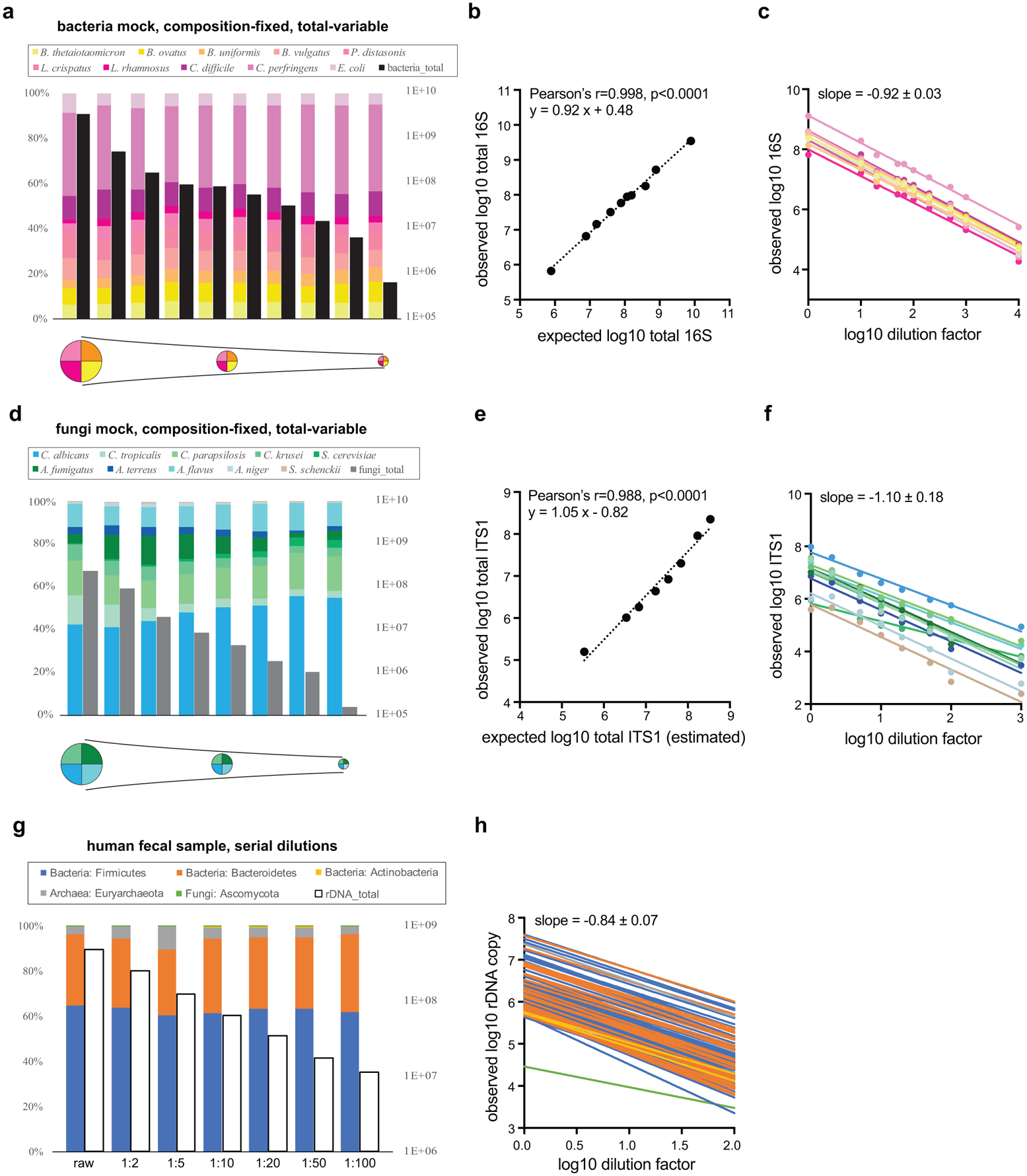
A set of single-kingdom mock communities with a fixed composition of 10 bacterial (a) or 10 fungal (d) species and variable total microbial loads (indicated by the pie chart schematics underneath), were quantified using MK-SpikeSeq for relative composition (colored bars) and absolute abundance (black/grey bars). b, e, Correlations between expected (based on initial microbial densities and known dilution factors) and MK-SpikeSeq-measured total absolute abundances show that MK-SpikeSeq reliably detects absolute abundances of bacteria and fungi. Note that for e, as exact rDNA copy numbers per fungal cell are undefined, the expected total ITS1 abundances are only estimates (here using 200 rDNA copies per fungal cell). c, f, Absolute abundance changes for individual members (color coded same as a, d) in the bacterial and fungal mock communities are largely consistent with known dilution factors. g, A set of serial dilutions of a human fecal sample was quantified using MK-SpikeSeq for relative composition (colored bars, shown are the phylum level taxa) and absolute abundance (empty bars). h, Absolute abundance changes for individual OTUs (color coded in phyla same as g) across kingdoms are largely consistent with known dilution factors.
