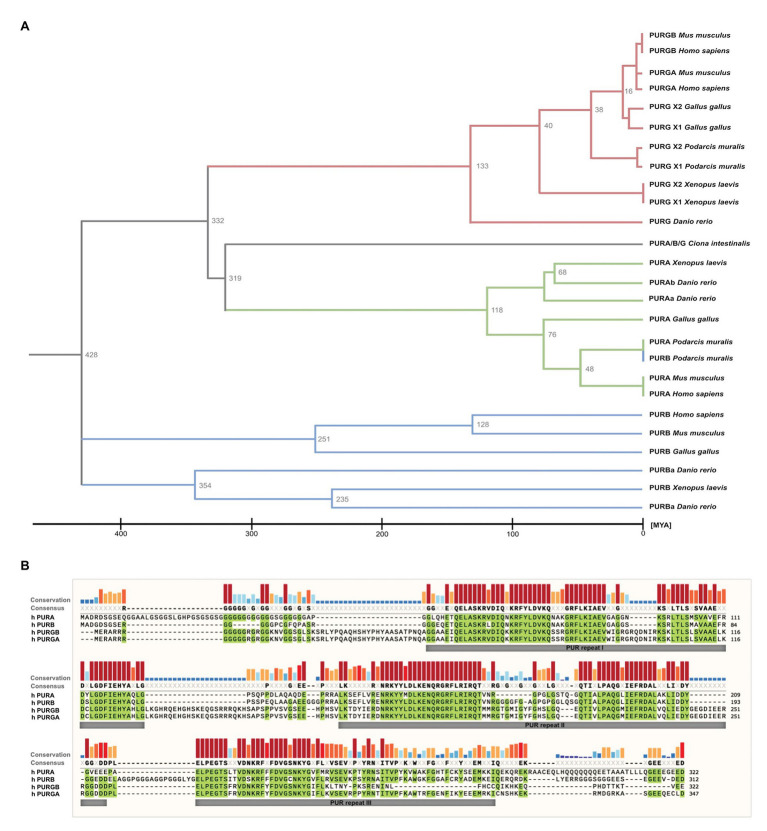
Figure 2.
Sequence similarities between PUR-family proteins. (A) Phylogenetic tree of the PUR-proteins of selected species. The tree shows evolutionary time on the X axis and indicates a differentiation of the PUR proteins between 428 and 319 Million Years Ago (MYA). The tree was inferred using the RelTime method and was computed using one calibration constraint. Branch lengths were calculated using the Maximum Likelihood (ML) method and the JTT matrix-based substitution model. A discrete Gamma distribution was used to model evolutionary rate differences among sites. The phylogenetic analysis was performed with the program MEGAX (Hall, 2013; Mello, 2018). (B) Amino acid sequence alignment of human PURA, PURB, PURG variant A, and PURG variant B. Sequence identity is highlighted in green. Whereas human PURA and PURB proteins share 70% sequence conservation, PURG has 48% identity to PURA and thus is more divergent. For all paralogs, sequence similarity is notably higher within the PUR repeats. The alignment was executed using the MUSCLE algorithm and program MEGAX.