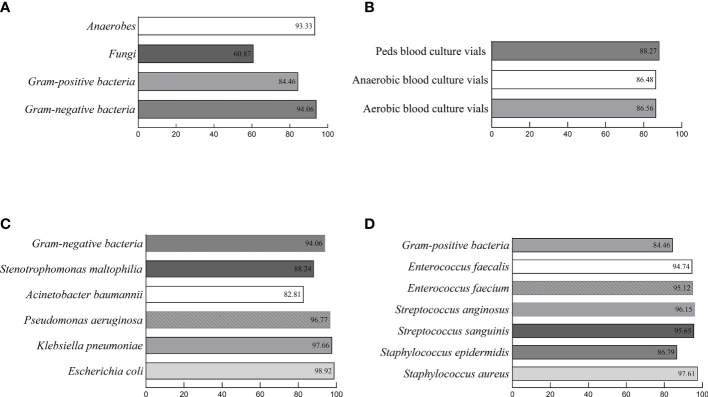
Figure 3.
Consistency rates of the optimized protocol for direct identification of microorganisms from positive blood cultures with the conventional method. (A). The percentages of concordant results were 93.33%/60.87%/84.46%/94.06%, respectively for Fungi, Anaerobes, Gram-positive bacteria, and Gram-negative bacteria with a log(score) of ≥1.700. (B). The percentages of concordant results were 88.27%/86.48%/86.56%, respectively for Peds/Anaerobic/Aerobic blood culture vials with a log(score) of ≥1.700. (C). The percentages of concordant results were 88.24%/82.81%/96.77%/97.66%/98.92%, respectively for general gram-negative bacteria Stenotrophomonas maltophilia, Acinetobacter baumannii, Pseudomonas aeruginosa, Klebsiella pneumonia, and Escherichia coli with a log(score) of ≥1.700. (D). The percentages of concordant results were 94.74%/95.12%/96.15%/95.65%/86.79%/97.61%, respectively for general gram- positive bacteria Enterococcus faecalis, Enterococcus faecium, Streptococcus anginosus, Streptococcus sanguini, Staphylococcus epidermidis, and Staphylococcus aureus with a log(score) of ≥1.700.