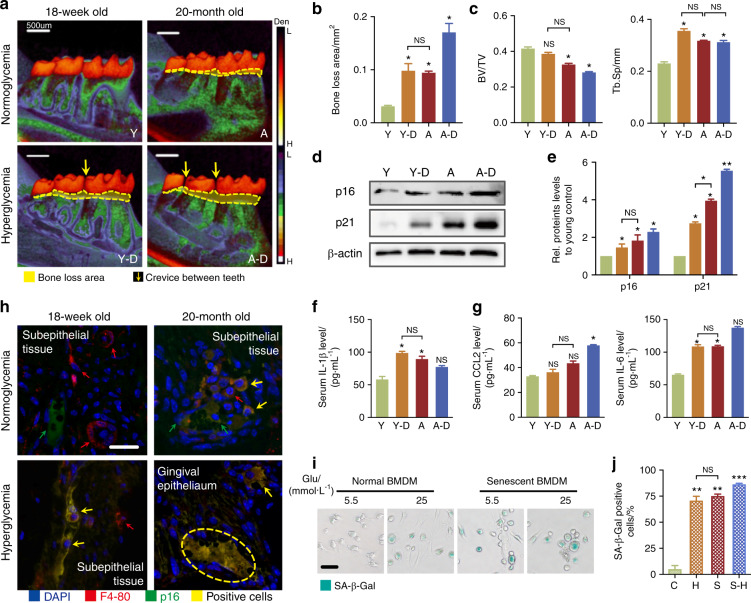
Fig. 1.
Activation of p16/p21 and SASP in periodontal lesions of diabetic and aged mice. a Periodontal damage in diabetic and aged mice. Micro-CT analysis showed the periodontal damage. The area of bone loss was shown by the area of density reduction below the crown. The alveolar bone resorption area is indicated by yellow line areas. Scale bar: 500 μm. b Quantification of alveolar bone resorption area (the area of density reduction below the crown). c Quantification of bone volume/total volume (BV/TV) and Tb.Sp (trabecular separation). d p16 and p21 were detected by western blot. e Protein p16/p21 levels relative to β‐actin protein levels were assessed by densitometric analysis and expressed as a percentage of the levels of wild‐type mice. f, g Serum early/late SASP response in diabetic and aged mice. The SASP responses (Supplementary Table 2) were detected by ELISA, including early SASP (IL-1β) and late SASP response (CCL2 and IL-6). h The location of p16+ macrophage in periodontal lesions. Immunohistochemistry staining was applied to co-localize p16 and F4-80 (macrophage mark) protein expression in periodontal tissues. Note the bright p16 flake in cells that co-localized with a prominent flake of F4-80. The yellow arrow indicated the double-positive cells. The red arrow indicated the F4-80+-positive cells and the green arrow indicated the p16+-positive cells. Scale bar = 20 μm. i, j Features and quantification of high-glucose-induced and drug-induced (DIS) senescent BMDMs. SA-β-Gal staining was used to display the senescent cells. The green represented activation of senescence. Scale bar: 50 μm. Y, young normoglycemic mice (18-wk-old); D, young-diabetic mice; A, aged normoglycemic mice (20-mo-old); A–D, aged-diabetic mice. C, BMDM cultured in low-glucose (5.5 mmol·L−1) condition; H, BMDM cultured in high-glucose (25 mmol·L−1) condition; S, DIS-BMDM cultured in low-glucose condition; SH, DIS-BMDM cultured in high-glucose condition. *P < 0.05. **P < 0.01. ***P < 0.001. NS, no significance. Repeated three times. Data are presented as the mean ± SD (n = 10) and compared to the normal