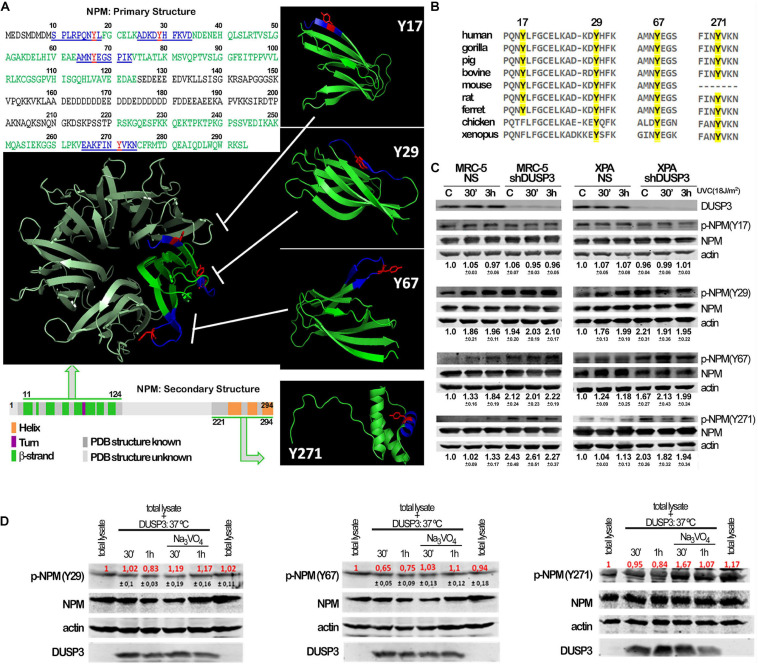
FIGURE 2.
Conserved tyrosine residues of NPM are dephosphorylated by DUSP3. (A) The complete amino acid sequence of NPM protein shows the four tyrosine residues (in red: Y17, Y29, Y67, and Y271) inserted in decapeptide sequences (in blue) within the primary structure of NPM. The four tyrosine are localized in two regions of the primary and secondary structures of NPM (in green: one at very end N-terminal and the other at very end C-terminal), interspaced by a structurally unknown region (in gray), which have been crystallized and better studied. The four Tyr-containing decapeptide sequences were used as template to synthesize four decaphosphopeptides phosphorylated on each specific Tyr residue, which were used to immunization of rabbits and generation of phospho-specific antibodies. The oligomeric NPM structure (pentameric) proposed from functional studies (in metallic green; PDB 5EHD) was used as model to highlight the spatial position of the four tyrosines on NPM 3D structure (in green; PDB, 5EHD, and 2VXD). (B) Interspecies multiple alignment of the regions containing the four NPM tyrosines show these residues conservation throughout evolution. (C) Cellular lysates from MRC-5 and XPA cell lines (NS or shDUSP3) exposed to 18 J/m2 UVC radiation were immunoblotted using the antibodies against the four phospho-tyrosine residues of NPM. DUSP3 knockdown increased the phosphorylation of the 29, 67, and 271 tyrosines. (D) The in vitro dephosphorylation assays confirmed that DUSP3 can specifically dephosphorylate three tyrosine residues (29, 67, and 271), since their phosphorylation levels are decreased by the addition of exogenous DUSP3 to the lysates but are restored in the presence of Na3VO4. The immunoblotting images are representative of three independent experiments and the quantification is shown around the bands as mean (red) ± standard deviation (black).