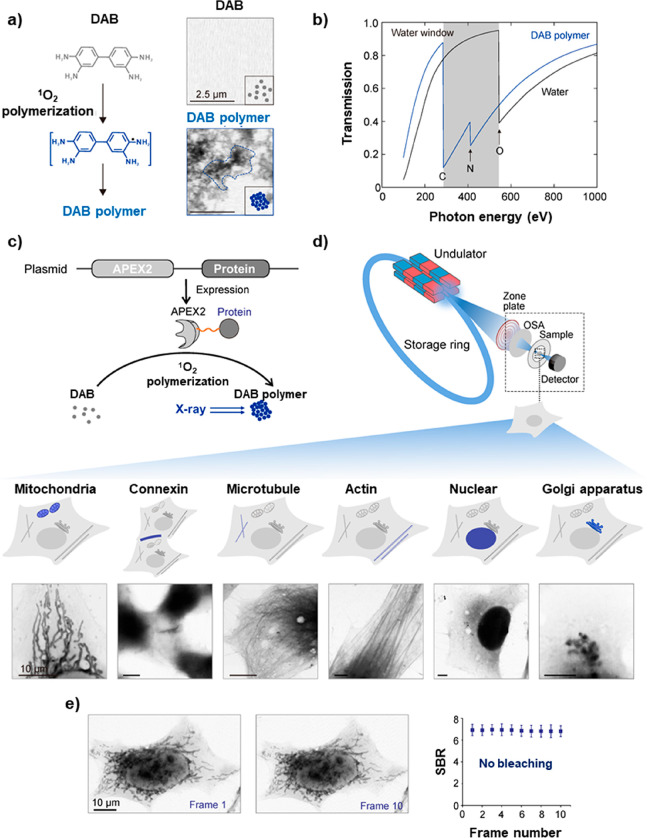
Figure 7.
Repurpose engineered peroxidase as genetically encoded tags for protein localization with XRM. (a) Schematics showing the catalytic polymerization of 3,3′-diaminobenzidine (DAB) into DAB polymer (left) and X-ray imaging of DAB polymer (right). (b) X-ray absorption spectra of water and DAB polymer. In the “water window”, absorption by carbon and nitrogen is much stronger than by oxygen. (c) Schematics showing APEX2 as a genetically encoded tag for protein localization with XRM. By using fusion expression plasmids including APEX2 and biotargets, these tags are highly specific and can polymerize DAB into localized X-ray-visible dense DAB polymers. This strategy enables localizing and imaging various cellular targets with high resolution. (d) STXM images of cellular proteins and specific amino acid sequences: COX4 (mitochondrial), Cx43, α-tubulin, β-actin, NLS, and GalT. Scale bars: 10 μm. (e) Photostability characterization of the genetically encoded tag for protein localization with XRM. No photobleaching occurred after 10 frames of STXM scans (for each STXM scan, the signal-to-background ratio of 10 loci was calculated and averaged to obtain a single value). Scale bars: 10 μm. Reprinted with permission from ref (230) under a CC-BY License. Copyright 2020 Oxford University Press.