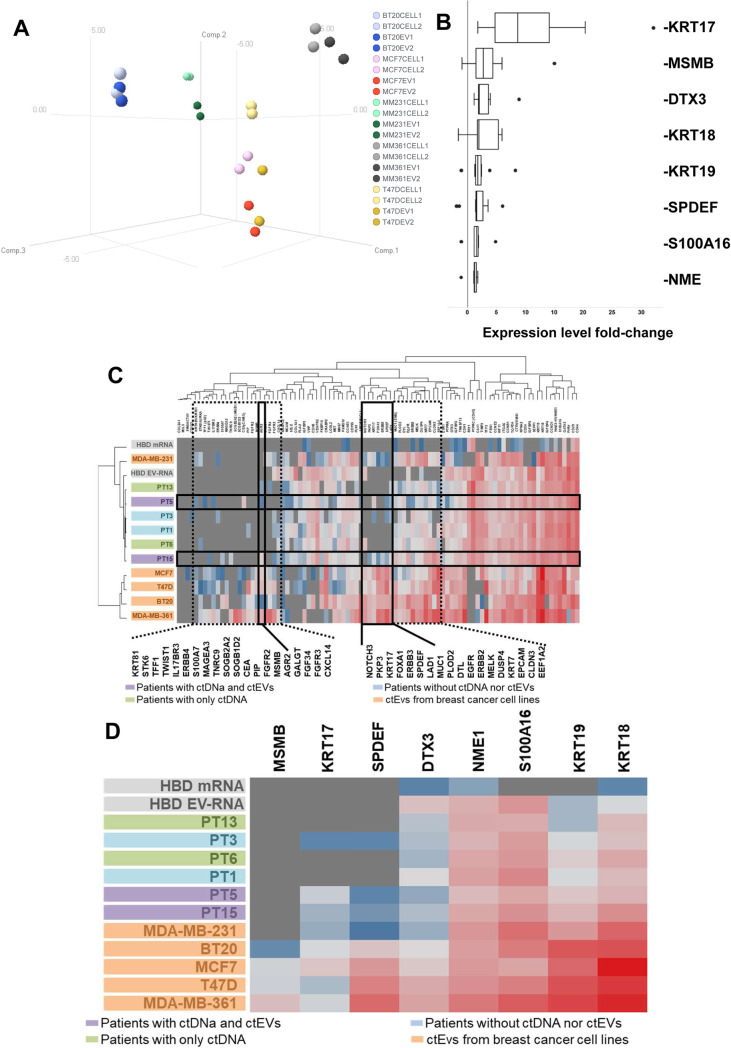
Fig. 4.
Gene expression profiling of EV-RNA. Expression of 96 genes was evaluated by RT-qPCR in EV-RNA and cellular mRNA from cancer cell lines and HBDs and EV-RNA from patients. a 3D Principal Component analysis plot of the 96 gene expression profiles in EVs and matching tumor cells from 5 breast cancer cell lines. Each dot represents a sample. Axes show the first three principal components. Dark colored dots indicate the EV-RNA samples and the lighter colors the cellular mRNA. b Enriched genes in EVs are indicated on the Y-axis while the X-axis shows the expression fold-change in EV-RNAs compared to their matched cellular mRNA from 5 breast cancer cell lines. Each boxplot consists of data from duplicate analysis of EV-RNA and tumor cell mRNA. c Hierarchical cluster analyses of EV-RNA expression profiles from patients, HBD and breast cancer cell lines compared to median HBD leucocyte mRNA expression. Clustering is shown for 96 genes (upper plot) and for the 8 genes enriched in tumor cell line EVs (lower plot; see also Fig. 2b). The boxes in the upper plot indicate genes upregulated in EVs from tumor cells compared to HBDs (dashed black boxes), and tumor cell related genes (solid black boxes) expressed in cell lines and in patients with ctEVs and ctDNA (PT5 and 15). Expression levels of target genes were compared to the average of reference genes HMBS, HPRT1 and GUSB levels; grey color indicates no expression, blue color is below reference gene level, and red color is above reference gene level