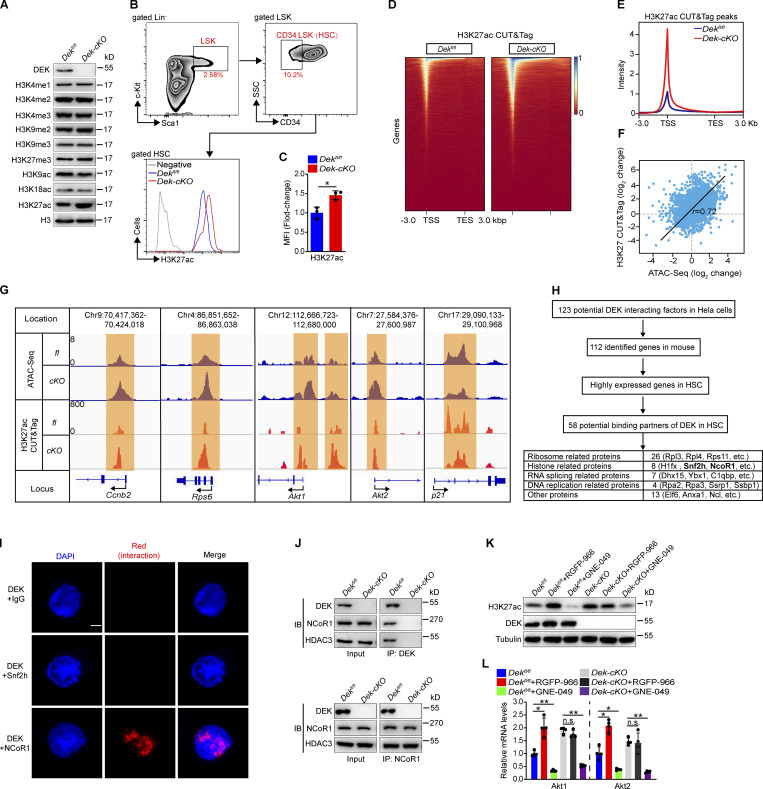
Figure 5.
DEK induces deacetylation of H3K27 by recruiting the corepressor NCoR1. (A) Western blot for modified histone 3 in lysates prepared from freshly sorted Lin−c-Kit+ cells. (B and C) FACS analysis of H3K27ac level in HSCs (CD34−LSK). The histograms indicate the mean fluorescence intensity (MFI) analysis of H3K27ac in HSCs (n = 3). (D) Representative heatmap of genome-wide H3K27ac CUT&Tag signal around genes. (E) Average diagram of genome-wide H3K27ac CUT&Tag peaks at TSS regions (±3,000 bp). (F) Correlation of changes between ATAC peaks and H3K27ac CUT&Tag peaks. Correlation coefficient (r) and P values (r = 0.72, P < 1 × 10−50) were calculated by Pearson’s correlation analysis. (G) Distribution of ATAC peaks and H3K27ac CUT&Tag peaks across the indicated gene loci. (H) Schematic representation of the workflow for DEK partners’ discovery. Potential DEK interacting factors from published mass spectrometry data (Smith et al., 2018). The highly expressed genes in HSCs have an FPKM value of >30 (data from RNA-seq). (I) In situ ligation assay to detect DEK/Snf2h and DEK/NCoR1 interaction. As a negative control, proximity ligation was performed using a rabbit anti-DEK antibody and a mouse IgG. Nuclei were visualized using DAPI staining. Scale bar: 5 µm. (J) Lin−c-Kit+ cells were freshly isolated from mice. DEK or NCoR1 protein was immunoprecipitated from cell lysates, followed by immunoblotting (IB). (K) Western blot for H3K27ac, DEK, and tubulin in lysates prepared from Lin−c-Kit+ cells. Lin−c-Kit+ cells were sorted and cultured in vitro, with treatment of RGFP-966 (5 µM) or GNE-049 (500 nM) for 24 h. (L) qRT-PCR analysis of the indicated transcripts from HSCs (n = 4). HSCs were sorted and cultured in vitro, with treatment with RGFP-966 (5 µM) or GNE-049 (500 nM) for 24 h. Error bars represent means ± SD. *, P < 0.05, **, P < 0.01; Student’s t test or one-way ANOVA. Data in A–C and I–L are representative of three independent experiments. TES, transcriptional end site.