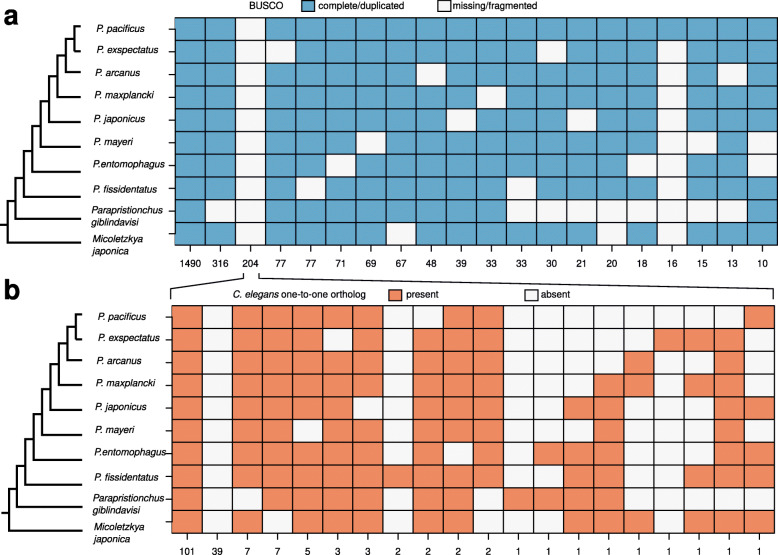
Fig. 3.
Missing BUSCO orthologs in diplogastrid genomes. a Classifications from the BUSCO results (nematode odb10) were summarized across all diplogastrid genomes and the twenty most abundant patterns are displayed. While misannotations should result in random patterns with low frequency, highly abundant patterns can point towards problematic data sets (e.g. Parapristionchus), true biological events (e.g. gene losses) or systematic biases. b Complementary ortholog predictions based on best-reciprocal BLASTP hits show that 101 (49.5%) of BUSCO genes, which were predicted to be missing in all diplogastrid genomes, have putative one-to-one orthologs