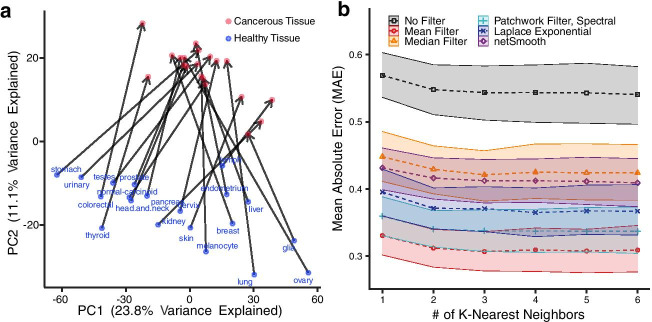
Fig. 3.
Denoising to predict protein expression changes in healthy and cancerous tissues. Tests of the network filters on a cancer protein expression prediction task. In this test, we predict the protein expression changes that occur when a healthy tissue becomes cancerous, quantified by the out-of-sample prediction accuracy with and without using network filters to preprocess the data before training. a The first two principal components of immunohistochemistry data of healthy and cancerous tissues in the Human Protein Atlas. Arrows connect a healthy tissue (blue) to the corresponding cancer (red). The first component captures variations across tissues, while the second captures variation in state (healthy vs. cancerous). Predicting the precise changes between healthy and cancerous tissues is a non-trivial task. Therefore, we perform a K-Nearest Neighbors regression on the HPA data, with and without preprocessing with network filters. We evaluate the model by leave-one-out cross validation, and calculating the MAE of the predicted and actual data values for the left out healthy-cancerous pair. b All network filters and diffusion methods improve the MAE compared to the no-filter baseline. We compare this across different choices of K, as it is a free parameter. The shaded areas represent 95% bootstrapped confidence intervals