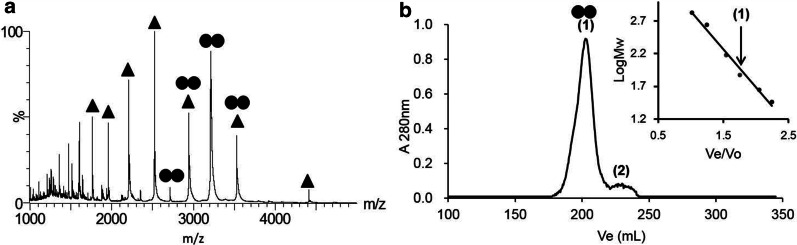
Fig. 4.
Oligomerization of CP12 revealed by native ESI–MS and size-exclusion chromatography. a ESI–MS spectrum of CP12. Signals labelled with a single triangle are assigned to monomeric CP12 (envelope of m/z 1765.2 to 4411.4 with charged states ranging from 10 to 4), and signals labelled with two circles are assigned to dimeric CP12 (envelope of m/z 2715.1 to 3529.4 with charged states ranging from 13 to 10). b Elution profile of CP12 on a S200 size-exclusion column. The single elution peak is assigned to dimeric CP12 (labelled with (1)). A small proportion of a smaller particle is assigned with the labelled (2) and is ascribed to impurities. The column was calibrated with proteins of known relative molecular mass (insert): Thyroglobulin (670 kDa), Ferritin (440 kDa), Alcohol dehydrogenase (150 kDa), Conalbumin (75 kDa), Ovalbumin (44 kDa), carbonic anhydrase (30 kDa). The linear fit (R2 = 0.98) indicates a dependency of the MW as a function of the elution volume (Ve) as: with Vo being the elution volume of blue dextran (void volume of 110.3 mL). The elution volume of CP12 is indicated with an arrow