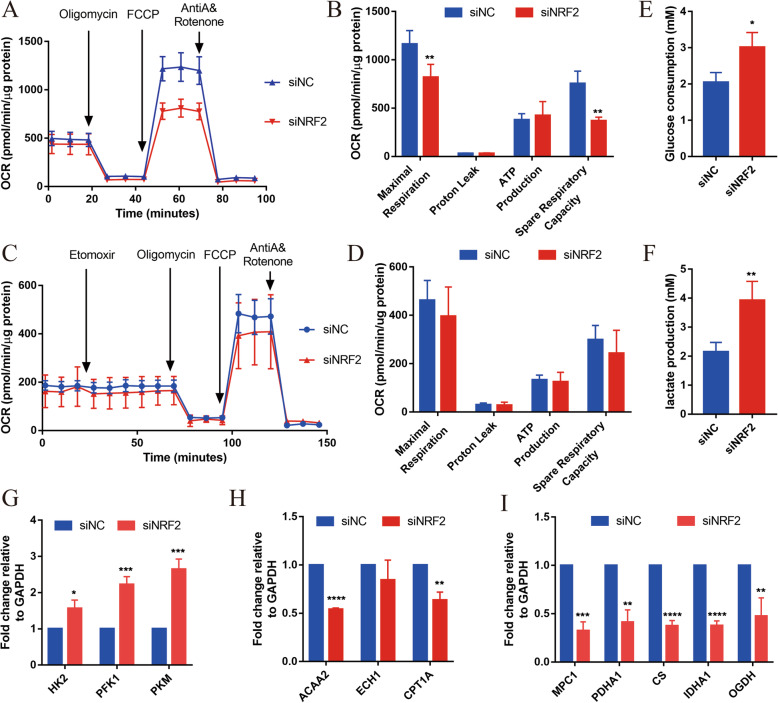
Fig. 3.
NRF2 is required for hiPSC-CM mitochondrial metabolism maturation. a Representative mitochondrial respiration in siNRF2 and siNC hiPSC-CMs after incubation with the ATP synthase inhibitor oligomycin, the respiratory uncoupler carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), and the respiratory chain blockers rotenone and antimycin A. b Quantification of maximal respiration capacity, proton leakage, ATP production, and spare respiratory capacity in siNRF2 and siNC hiPSC-CMs (n = 4 for each group). c Representative fatty acid oxidation in siNRF2 and siNC hiPSC-CMs after incubation with the specific inhibitor of carnitine palmitoyl transferase 1A (CPT1A) Etomoxir, oligomycin, FCCP, and rotenone and antimycin A. d Quantification of maximal respiration capacity, proton leakage, ATP production, and spare respiratory capacity in siNRF2 and siNC hiPSC-CMs (n = 4 for each group). The means ± SEM are shown. **p < 0.01. Glucose consumption (e) and lactic acid generation (f) of siNRF2 and siNC hiPSC-CMs were detected by kit. n = 3. Relative expression levels of the key metabolic genes in glycolysis (g), fatty acid β-oxidation (h), and aerobic oxidation (i). n = 3. The means ± SEM are shown. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001